1
Project-Euler/.../Project-Euler
ARCHIVE10
Project-Euler/.../Project-Euler
ARCHIVE11
Project-Euler/.../Project-Euler
ARCHIVE12
Project-Euler/.../Project-Euler
ARCHIVE13
Project-Euler/.../Project-Euler
ARCHIVE14
Project-Euler/.../Project-Euler
ARCHIVE15
Project-Euler/.../Project-Euler
ARCHIVE16
Project-Euler/.../Project-Euler
ARCHIVE17
Project-Euler/.../Project-Euler
ARCHIVE18
Project-Euler/.../Project-Euler
ARCHIVE19
Project-Euler/.../Project-Euler
ARCHIVE2
Project-Euler/.../Project-Euler
ARCHIVE20
Project-Euler/.../Project-Euler
ARCHIVE2020-Fall
The…/.../The-Chinese-University-of-Hong-Kong-Shenzhen
ARCHIVE2021-Fall
The…/.../The-Chinese-University-of-Hong-Kong-Shenzhen
ARCHIVE2021-Spring
The…/.../The-Chinese-University-of-Hong-Kong-Shenzhen
ARCHIVE2021-Summer
The…/.../The-Chinese-University-of-Hong-Kong-Shenzhen
ARCHIVE2022-Fall
The…/.../The-Chinese-University-of-Hong-Kong-Shenzhen
ARCHIVE2022-Spring
The…/.../The-Chinese-University-of-Hong-Kong-Shenzhen
ARCHIVE2023-Fall
The…/.../The-Chinese-University-of-Hong-Kong-Shenzhen
ARCHIVE2023-Spring
The…/.../The-Chinese-University-of-Hong-Kong-Shenzhen
ARCHIVE2024-Spring
The…/.../The-Chinese-University-of-Hong-Kong-Shenzhen
ARCHIVE20K-smooth
The-Chinese-University-of-Hong-Ko…/.../scripts
ARCHIVE20K-smooth (no image)
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVE21
Project-Euler/.../Project-Euler
ARCHIVE22
Project-Euler/.../Project-Euler
ARCHIVE23
Project-Euler/.../Project-Euler
ARCHIVE24
Project-Euler/.../Project-Euler
ARCHIVE25
Project-Euler/.../Project-Euler
ARCHIVE26
Project-Euler/.../Project-Euler
ARCHIVE27
Project-Euler/.../Project-Euler
ARCHIVE28
Project-Euler/.../Project-Euler
ARCHIVE29
Project-Euler/.../Project-Euler
ARCHIVE3
Project-Euler/.../Project-Euler
ARCHIVE30
Project-Euler/.../Project-Euler
ARCHIVE31
Project-Euler/.../Project-Euler
ARCHIVE32
Project-Euler/.../Project-Euler
ARCHIVE33
Project-Euler/.../Project-Euler
ARCHIVE34
Project-Euler/.../Project-Euler
ARCHIVE35
Project-Euler/.../Project-Euler
ARCHIVE36
Project-Euler/.../Project-Euler
ARCHIVE37
Project-Euler/.../Project-Euler
ARCHIVE4
Project-Euler/.../Project-Euler
ARCHIVE5
Project-Euler/.../Project-Euler
ARCHIVE6
Project-Euler/.../Project-Euler
ARCHIVE7
Project-Euler/.../Project-Euler
ARCHIVE8
Project-Euler/.../Project-Euler
ARCHIVE9
Project-Euler/.../Project-Euler
ARCHIVEappendix
The-Chinese-University-…/.../CSC4005-Project-1
ARCHIVEappendix
The-Chinese-University-…/.../CSC4005-Project-2
ARCHIVEappendix
The-Chinese-University-…/.../CSC4005-Project-4
ARCHIVEarchive
ARCHIVEBIO1001-General-Biology
The-Chinese-University-of-Hon…/.../2021-Summer
ARCHIVEbonus
The-Chinese-Universi…/.../CSC3150-Assignment-2
ARCHIVEbonus
The-Chinese-Universi…/.../CSC3150-Assignment-3
ARCHIVEbucketsort
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVECHM1001-General-Chemistry
The-Chinese-University-of-Hong-…/.../2020-Fall
ARCHIVEcollections
The-Chinese-University-of-…/.../StanfordCPPLib
ARCHIVECompetitive-Programming
ARCHIVEcomponents
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEcpu
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVECSC1001-Introduction-to-Computer-Science-Programming-Methodology
The-Chinese-University-of-Hon…/.../2021-Spring
ARCHIVECSC1002-Computational-Laboratory
The-Chinese-University-of-Hon…/.../2021-Spring
ARCHIVECSC3001-Discrete-Mathematics
The-Chinese-University-of-Hong-…/.../2021-Fall
ARCHIVECSC3002-Assignment-1-src
The…/.../CSC3002-Introduction-to-Computer-Science-Programming-Paradigms
ARCHIVECSC3002-Assignment-2-src
The…/.../CSC3002-Introduction-to-Computer-Science-Programming-Paradigms
ARCHIVECSC3002-Assignment-3-src
The…/.../CSC3002-Introduction-to-Computer-Science-Programming-Paradigms
ARCHIVECSC3002-Assignment-4-src
The…/.../CSC3002-Introduction-to-Computer-Science-Programming-Paradigms
ARCHIVECSC3002-Assignment-5-src
The…/.../CSC3002-Introduction-to-Computer-Science-Programming-Paradigms
ARCHIVECSC3002-Assignment-6-src
The…/.../CSC3002-Introduction-to-Computer-Science-Programming-Paradigms
ARCHIVECSC3002-Introduction-to-Computer-Science-Programming-Paradigms
The-Chinese-University-of-Hon…/.../2022-Spring
ARCHIVECSC3050-Computer-Architecture
The-Chinese-University-of-Hon…/.../2022-Spring
ARCHIVECSC3050-Project-1-assembler
The-Chinese…/.../CSC3050-Computer-Architecture
ARCHIVECSC3050-Project-2-simulator
The-Chinese…/.../CSC3050-Computer-Architecture
ARCHIVECSC3050-Project-3-verilog
The-Chinese…/.../CSC3050-Computer-Architecture
ARCHIVECSC3050-Project-4-CPU
The-Chinese…/.../CSC3050-Computer-Architecture
ARCHIVECSC3100-Data-Structures
The-Chinese-University-of-Hong-…/.../2022-Fall
ARCHIVECSC3150-Assignment-1
The-Chinese-Uni…/.../CSC3150-Operating-Systems
ARCHIVECSC3150-Assignment-2
The-Chinese-Uni…/.../CSC3150-Operating-Systems
ARCHIVECSC3150-Assignment-3
The-Chinese-Uni…/.../CSC3150-Operating-Systems
ARCHIVECSC3150-Assignment-4
The-Chinese-Uni…/.../CSC3150-Operating-Systems
ARCHIVECSC3150-Operating-Systems
The-Chinese-University-of-Hong-…/.../2022-Fall
ARCHIVECSC3170-Assignment-2
The-Chinese-Unive…/.../CSC3170-Database-System
ARCHIVECSC3170-Database-System
The-Chinese-University-of-Hon…/.../2023-Spring
ARCHIVECSC4001-Project-1
The-Chinese-…/.../CSC4001-Software-Engineering
ARCHIVECSC4001-Software-Engineering
The-Chinese-University-of-Hon…/.../2023-Spring
ARCHIVECSC4005-Parallel-Programming
The-Chinese-University-of-Hong-…/.../2023-Fall
ARCHIVECSC4005-Project-1
The-Chinese-…/.../CSC4005-Parallel-Programming
ARCHIVECSC4005-Project-2
The-Chinese-…/.../CSC4005-Parallel-Programming
ARCHIVECSC4005-Project-3
The-Chinese-…/.../CSC4005-Parallel-Programming
ARCHIVECSC4005-Project-4
The-Chinese-…/.../CSC4005-Parallel-Programming
ARCHIVECSC4008-Assignment-1
The-Ch…/.../CSC4008-Techniques-for-Data-Mining
ARCHIVECSC4008-Assignment-2
The-Ch…/.../CSC4008-Techniques-for-Data-Mining
ARCHIVECSC4008-Assignment-3
The-Ch…/.../CSC4008-Techniques-for-Data-Mining
ARCHIVECSC4008-Assignment-4
The-Ch…/.../CSC4008-Techniques-for-Data-Mining
ARCHIVECSC4008-Techniques-for-Data-Mining
The-Chinese-University-of-Hong-…/.../2023-Fall
ARCHIVECSC4120-Design-and-Analysis-of-Algorithms
The-Chinese-University-of-Hon…/.../2024-Spring
ARCHIVECSC4130-Assignment-1
The…/.../CSC4130-Introduction-to-Human-Computer-Interaction
ARCHIVECSC4130-Assignment-2
The…/.../CSC4130-Introduction-to-Human-Computer-Interaction
ARCHIVECSC4130-Assignment-3
The…/.../CSC4130-Introduction-to-Human-Computer-Interaction
ARCHIVECSC4130-Assignment-4
The…/.../CSC4130-Introduction-to-Human-Computer-Interaction
ARCHIVECSC4130-Introduction-to-Human-Computer-Interaction
The-Chinese-University-of-Hong-…/.../2022-Fall
ARCHIVEcss
The-Chinese-Universi…/.../DDA2003-Assignment-1
ARCHIVEcss
The-Chinese-Universi…/.../DDA2003-Assignment-3
ARCHIVEdata
The-Chinese-Universi…/.../CSC4130-Assignment-3
ARCHIVEdata
The-Chinese-Universi…/.../CSC4008-Assignment-2
ARCHIVEdata
The-Chinese-Universi…/.../CSC4008-Assignment-3
ARCHIVEdata
The-Chinese-Universi…/.../DDA2003-Assignment-1
ARCHIVEdata
The-Chinese-Universi…/.../DDA2003-Assignment-2
ARCHIVEdata
The-Chinese-Universi…/.../DDA2003-Assignment-3
ARCHIVEdata
The-Chinese-Universi…/.../DDA2003-Assignment-4
ARCHIVEdata
The-Chinese-Universi…/.../DDA2003-Assignment-5
ARCHIVEdata-Q1
The-Chinese-Universi…/.../CSC4008-Assignment-1
ARCHIVEdata-Q1
The-Chinese-Universi…/.../CSC4008-Assignment-4
ARCHIVEdata-Q2
The-Chinese-Universi…/.../CSC4008-Assignment-1
ARCHIVEdata-Q2
The-Chinese-Universi…/.../CSC4008-Assignment-4
ARCHIVEDDA2003-Assignment-1
The-Chinese-Univ…/.../DDA2003-Visual-Analytics
ARCHIVEDDA2003-Assignment-2
The-Chinese-Univ…/.../DDA2003-Visual-Analytics
ARCHIVEDDA2003-Assignment-3
The-Chinese-Univ…/.../DDA2003-Visual-Analytics
ARCHIVEDDA2003-Assignment-4
The-Chinese-Univ…/.../DDA2003-Visual-Analytics
ARCHIVEDDA2003-Assignment-5
The-Chinese-Univ…/.../DDA2003-Visual-Analytics
ARCHIVEDDA2003-Assignment-6
The-Chinese-Univ…/.../DDA2003-Visual-Analytics
ARCHIVEDDA2003-Visual-Analytics
The-Chinese-University-of-Hon…/.../2023-Spring
ARCHIVEDDA3020-Assignment-1
The-Chinese-Univ…/.../DDA3020-Machine-Learning
ARCHIVEDDA3020-Assignment-2
The-Chinese-Univ…/.../DDA3020-Machine-Learning
ARCHIVEDDA3020-Assignment-3
The-Chinese-Univ…/.../DDA3020-Machine-Learning
ARCHIVEDDA3020-Assignment-4
The-Chinese-Univ…/.../DDA3020-Machine-Learning
ARCHIVEDDA3020-Machine-Learning
The-Chinese-University-of-Hong-…/.../2022-Fall
ARCHIVEDDA4210-Advanced-Machine-Learning
The-Chinese-University-of-Hon…/.../2024-Spring
ARCHIVEDDA6050-Analysis-of-Algorithms
The-Chinese-University-of-Hong-…/.../2023-Fall
ARCHIVEDDA6050-Assignment-2
The-Chines…/.../DDA6050-Analysis-of-Algorithms
ARCHIVEDDA6050-Assignment-4
The-Chines…/.../DDA6050-Analysis-of-Algorithms
ARCHIVEDDA6205-Game-Theory-and-Its-Applications
The-Chinese-University-of-Hon…/.../2024-Spring
ARCHIVEEIE2050-Digital-Logics-and-Systems
The-Chinese-University-of-Hong-…/.../2021-Fall
ARCHIVEgpu
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEgpu
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEgraphics
The-Chinese-University-of-…/.../StanfordCPPLib
ARCHIVEGray
The-Chinese-University-of-Hong-Ko…/.../scripts
ARCHIVEGray (no image)
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEinputs
The-Chinese-University-of-…/.../party-together
ARCHIVEio
The-Chinese-University-of-…/.../StanfordCPPLib
ARCHIVEjs
The-Chinese-Universi…/.../DDA2003-Assignment-1
ARCHIVEjs
The-Chinese-Universi…/.../DDA2003-Assignment-3
ARCHIVELena-smooth
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVELena-smooth
The-Chinese-University-of-Hong-Ko…/.../scripts
ARCHIVElib
The…/.../CSC3002-Introduction-to-Computer-Science-Programming-Paradigms
ARCHIVEMAT1001-Calculus-I
The-Chinese-University-of-Hong-…/.../2020-Fall
ARCHIVEMAT1002-Calculus-II
The-Chinese-University-of-Hon…/.../2021-Spring
ARCHIVEMAT2040-Linear-Algebra
The-Chinese-University-of-Hong-…/.../2021-Fall
ARCHIVEMAT3007-Optimization
The-Chinese-University-of-Hong-…/.../2023-Fall
ARCHIVEmatrices
The-Chinese-University-of-Hong-K…/.../project2
ARCHIVEmergesort
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEodd-even-sort
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEoutputs
The-Chinese-University-of-…/.../party-together
ARCHIVEparty-together
The…/.../CSC4120-Design-and-Analysis-of-Algorithms
ARCHIVEPHY1001-Mechanics
The-Chinese-University-of-Hong-…/.../2020-Fall
ARCHIVEPHY1002-Physics-Laboratory
The-Chinese-University-of-Hon…/.../2023-Spring
ARCHIVEpretest1
The-Chinese-University-…/.../CSC4001-Project-1
ARCHIVEpretest2
The-Chinese-University-…/.../CSC4001-Project-1
ARCHIVEprivate
The-Chinese-University-of-…/.../StanfordCPPLib
ARCHIVEprofiling
The-Chinese-University-…/.../CSC4005-Project-3
ARCHIVEprofiling
The-Chinese-University-…/.../CSC4005-Project-4
ARCHIVEprogram1
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVEprogram2
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVEProject-Euler
ARCHIVEproject1
The-Chinese-University-…/.../CSC4005-Project-1
ARCHIVEproject2
The-Chinese-University-…/.../CSC4005-Project-2
ARCHIVEquicksort
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEresults
The-Chinese-Universi…/.../DDA3020-Assignment-1
ARCHIVEresults
The-Chinese-University-…/.../CSC4005-Project-1
ARCHIVESchOJ
The…/.../The-Chinese-University-of-Hong-Kong-Shenzhen
ARCHIVEscripts
The-Chinese-University-…/.../CSC4005-Project-1
ARCHIVEscripts
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEsource
The-Chinese-Universi…/.../CSC3150-Assignment-1
ARCHIVEsource
The-Chinese-Universi…/.../CSC3150-Assignment-2
ARCHIVEsource
The-Chinese-Universi…/.../CSC3150-Assignment-3
ARCHIVEsource
The-Chinese-Universi…/.../CSC3150-Assignment-4
ARCHIVEsrc
The-Chinese-Universi…/.../CSC4130-Assignment-1
ARCHIVEsrc
The-Chinese-Universi…/.../CSC4130-Assignment-2
ARCHIVEsrc
The-Chinese-Universi…/.../CSC4130-Assignment-3
ARCHIVEsrc
The-Chinese-Universi…/.../CSC4130-Assignment-4
ARCHIVEsrc
The-Chinese-University-…/.../CSC4005-Project-3
ARCHIVEsrc
The-Chinese-University-…/.../CSC4005-Project-4
ARCHIVEsrc
The-Chinese-Universi…/.../CSC3170-Assignment-2
ARCHIVEsrc
The-Chinese-University-…/.../CSC4001-Project-1
ARCHIVEsrc
The-Chinese-Universi…/.../DDA2003-Assignment-2
ARCHIVEsrc
The-Chinese-Universi…/.../DDA2003-Assignment-4
ARCHIVEsrc
The-Chinese-Universi…/.../DDA2003-Assignment-5
ARCHIVEsrc
The-Chinese-University-of-Hong-K…/.../project1
ARCHIVEsrc
The-Chinese-University-of-Hong-K…/.../project2
ARCHIVESTA2001-Probability-and-Statistics-I
The-Chinese-University-of-Hon…/.../2021-Spring
ARCHIVEStanfordCPPLib
The-Chinese-University-of-Hong-Kong-S…/.../lib
ARCHIVEsystem
The-Chinese-University-of-…/.../StanfordCPPLib
ARCHIVEtest_cases
The-Chinese-Universi…/.../CSC3170-Assignment-2
ARCHIVEtext-based-image-generation-for-cuhksz-buildings
The-Chi…/.../DDA4210-Advanced-Machine-Learning
ARCHIVEThe-Chinese-University-of-Hong-Kong-Shenzhen
ARCHIVEthread_pool
The-Chinese-University-of-Hong-Kong…/.../bonus
ARCHIVEutil
The-Chinese-University-of-…/.../StanfordCPPLib
ARCHIVE.clang-format
The-Chinese-University-…/.../CSC4005-Project-3
ARCHIVE.clang-format
The-Chinese-University-of-Hong-K…/.../project2
ARCHIVE.README
The-Chinese-Universi…/.../CSC4130-Assignment-4
ARCHIVE.README
The-Chinese-University-of-Hong-Kong…/.../bonus
ARCHIVE~$data.xlsx
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVE0022_names.txt
Project-Euler/.../22
ARCHIVE1.in
The-Chinese-University-of-Hong-Kon…/.../inputs
ARCHIVE1.out
The-Chinese-University-of-Hong-Ko…/.../outputs
ARCHIVE1.py
The-Chinese-University-of-Hong-K…/.../pretest1
ARCHIVE1.py
The-Chinese-University-of-Hong-K…/.../pretest2
ARCHIVE1.py
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVE10.in
The-Chinese-University-of-Hong-Kon…/.../inputs
ARCHIVE10.out
The-Chinese-University-of-Hong-Ko…/.../outputs
ARCHIVE10.xlsx
The-Chinese-University-of-Hong…/.../test_cases
ARCHIVE11.xlsx
The-Chinese-University-of-Hong…/.../test_cases
ARCHIVE12.xlsx
The-Chinese-University-of-Hong…/.../test_cases
ARCHIVE13.xlsx
The-Chinese-University-of-Hong…/.../test_cases
ARCHIVE14.xlsx
The-Chinese-University-of-Hong…/.../test_cases
ARCHIVE15.xlsx
The-Chinese-University-of-Hong…/.../test_cases
ARCHIVE16.xlsx
The-Chinese-University-of-Hong…/.../test_cases
ARCHIVE2.in
The-Chinese-University-of-Hong-Kon…/.../inputs
ARCHIVE2.out
The-Chinese-University-of-Hong-Ko…/.../outputs
ARCHIVE2.py
The-Chinese-University-of-Hong-K…/.../pretest1
ARCHIVE2.py
The-Chinese-University-of-Hong-K…/.../pretest2
ARCHIVE2.py
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVE2.xlsx
The-Chinese-University-of-Hong…/.../test_cases
ARCHIVE20_03.in
The-Chinese-University-of-Hong-Kon…/.../inputs
ARCHIVE20_03.out
The-Chinese-University-of-Hong-Ko…/.../outputs
ARCHIVE20_10.in
The-Chinese-University-of-Hong-Kon…/.../inputs
ARCHIVE20_10.out
The-Chinese-University-of-Hong-Ko…/.../outputs
ARCHIVE20K-parallel-benchmarking.png
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVE20K-parallel-benchmarking.xlsx
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVE3.in
The-Chinese-University-of-Hong-Kon…/.../inputs
ARCHIVE3.out
The-Chinese-University-of-Hong-Ko…/.../outputs
ARCHIVE3.py
The-Chinese-University-of-Hong-K…/.../pretest1
ARCHIVE3.py
The-Chinese-University-of-Hong-K…/.../pretest2
ARCHIVE3.xlsx
The-Chinese-University-of-Hong…/.../test_cases
ARCHIVE4.in
The-Chinese-University-of-Hong-Kon…/.../inputs
ARCHIVE4.out
The-Chinese-University-of-Hong-Ko…/.../outputs
ARCHIVE4.py
The-Chinese-University-of-Hong-K…/.../pretest1
ARCHIVE4.py
The-Chinese-University-of-Hong-K…/.../pretest2
ARCHIVE4.xlsx
The-Chinese-University-of-Hong…/.../test_cases
ARCHIVE40_03.in
The-Chinese-University-of-Hong-Kon…/.../inputs
ARCHIVE40_03.out
The-Chinese-University-of-Hong-Ko…/.../outputs
ARCHIVE40_10.in
The-Chinese-University-of-Hong-Kon…/.../inputs
ARCHIVE40_10.out
The-Chinese-University-of-Hong-Ko…/.../outputs
ARCHIVE404.html
The-Chinese-University-of-Hon…/.../thread_pool
ARCHIVE5.in
The-Chinese-University-of-Hong-Kon…/.../inputs
ARCHIVE5.out
The-Chinese-University-of-Hong-Ko…/.../outputs
ARCHIVE5.py
The-Chinese-University-of-Hong-K…/.../pretest1
ARCHIVE5.py
The-Chinese-University-of-Hong-K…/.../pretest2
ARCHIVE5.xlsx
The-Chinese-University-of-Hong…/.../test_cases
ARCHIVE6.in
The-Chinese-University-of-Hong-Kon…/.../inputs
ARCHIVE6.out
The-Chinese-University-of-Hong-Ko…/.../outputs
ARCHIVE6.py
The-Chinese-University-of-Hong-K…/.../pretest1
ARCHIVE6.py
The-Chinese-University-of-Hong-K…/.../pretest2
ARCHIVE6.xlsx
The-Chinese-University-of-Hong…/.../test_cases
ARCHIVE7_order.xlsx
The-Chinese-University-of-Hong…/.../test_cases
ARCHIVE7.in
The-Chinese-University-of-Hong-Kon…/.../inputs
ARCHIVE7.out
The-Chinese-University-of-Hong-Ko…/.../outputs
ARCHIVE7.py
The-Chinese-University-of-Hong-K…/.../pretest1
ARCHIVE7.py
The-Chinese-University-of-Hong-K…/.../pretest2
ARCHIVE8.in
The-Chinese-University-of-Hong-Kon…/.../inputs
ARCHIVE8.out
The-Chinese-University-of-Hong-Ko…/.../outputs
ARCHIVE8.xlsx
The-Chinese-University-of-Hong…/.../test_cases
ARCHIVE9.in
The-Chinese-University-of-Hong-Kon…/.../inputs
ARCHIVE9.out
The-Chinese-University-of-Hong-Ko…/.../outputs
ARCHIVE9.xlsx
The-Chinese-University-of-Hong…/.../test_cases
ARCHIVEa.cpp
The-Chinese-Universi…/.../DDA6050-Assignment-2
ARCHIVEa.cpp
The-Chinese-Universi…/.../DDA6050-Assignment-4
ARCHIVEA1_Data.csv
The-Chinese-University-of-Hong-Kong-…/.../data
ARCHIVEabort.c
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVEacndata_sessions.json
The-Chinese-University-of-Hong-Kong-…/.../data
ARCHIVEaddr2line.exe
The-Chinese-University-of-Hong-Kong-S…/.../lib
ARCHIVEaddr2line64.exe
The-Chinese-University-of-Hong-Kong-S…/.../lib
ARCHIVEalarm.c
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVEALU.v
The-Chinese-Uni…/.../CSC3050-Project-3-verilog
ARCHIVEApp.css
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEApp.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEapp.py
The…/.../text-based-image-generation-for-cuhksz-buildings
ARCHIVEApp.test.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEAssignment1.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-1-src
ARCHIVEAssignment1.h
The-Chinese-Univ…/.../CSC3002-Assignment-1-src
ARCHIVEassignment2.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-2-src
ARCHIVEassignment2.h
The-Chinese-Univ…/.../CSC3002-Assignment-2-src
ARCHIVEassignment3.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-3-src
ARCHIVEassignment3.h
The-Chinese-Univ…/.../CSC3002-Assignment-3-src
ARCHIVEassignment4.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-4-src
ARCHIVEassignment4.h
The-Chinese-Univ…/.../CSC3002-Assignment-4-src
ARCHIVEassignment5.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-5-src
ARCHIVEassignment5.h
The-Chinese-Univ…/.../CSC3002-Assignment-5-src
ARCHIVEAssignment6.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-6-src
ARCHIVEasync.c
The-Chinese-University-of-Hon…/.../thread_pool
ARCHIVEasync.h
The-Chinese-University-of-Hon…/.../thread_pool
ARCHIVEasync.o
The-Chinese-University-of-Hon…/.../thread_pool
ARCHIVEAtCoder - abc052a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc052b.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc053a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc053b.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc054a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc054c.cpp
Competitive-Progr��…/.../Competitive-Programming
ARCHIVEAtCoder - abc055a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc055b.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc056a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc056b.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc148a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc148b.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc148c.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc148d.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc148e.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc167a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc167b.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc167c.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc167d.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc170a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc170b.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc170c.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc170d.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc170e.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc272a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc272b.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc272c.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc272d.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc275a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc275b.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc275c.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc275d.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc275e.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc284a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc284b.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc284c.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc284d.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc284e.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc285a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc285b.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc285c.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc285d.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc285e.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc285f.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc287a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc287b.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc287c.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc287d.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc287e.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc287g.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc288a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc288b.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc288c.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc288d.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc295a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc295b.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc295c.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - abc295d.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - agc033a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - arc068a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - arc070a.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEAtCoder - arc085c.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEb.cpp
The-Chinese-Universi…/.../DDA6050-Assignment-2
ARCHIVEb.cpp
The-Chinese-Universi…/.../DDA6050-Assignment-4
ARCHIVEBanishLetters.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-1-src
ARCHIVEBanishLetters.h
The-Chinese-Univ…/.../CSC3002-Assignment-1-src
ARCHIVEBarChart.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEBarChart.js
The-Chinese-University-of-Hong…/.../components
ARCHIVEbase64.h
The-Chinese-University-of-Hong-Kong-Sh…/.../io
ARCHIVEbasicgraph.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEbigfloat.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEbigint.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-5-src
ARCHIVEbigint.h
The-Chinese-Univ…/.../CSC3002-Assignment-5-src
ARCHIVEbiginteger.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEBIO1001 - Homework 1.pdf
The-Chinese-Unive…/.../BIO1001-General-Biology
ARCHIVEBIO1001 - Homework 2.pdf
The-Chinese-Unive…/.../BIO1001-General-Biology
ARCHIVEBIO1001.grades
The-Chinese-Unive…/.../BIO1001-General-Biology
ARCHIVEbitstream.h
The-Chinese-University-of-Hong-Kong-Sh…/.../io
ARCHIVEbrowsing.txt
The-Chinese-University-of-Hong-Kong-…/.../data
ARCHIVEbuffer.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-4-src
ARCHIVEbuffer.h
The-Chinese-Univ…/.../CSC3002-Assignment-4-src
ARCHIVEbus.c
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVEc.cpp
The-Chinese-Universi…/.../DDA6050-Assignment-2
ARCHIVEc.cpp
The-Chinese-Universi…/.../DDA6050-Assignment-4
ARCHIVEc1.txt
The-Chinese-University-of-Hong-Ko…/.../data-Q1
ARCHIVEc2.txt
The-Chinese-University-of-Hong-Ko…/.../data-Q1
ARCHIVEcalendar.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-2-src
ARCHIVEcalendar.h
The-Chinese-Univ…/.../CSC3002-Assignment-2-src
ARCHIVEcall_stack.h
The-Chinese-University-of-Hong-Kon…/.../system
ARCHIVEcf888b.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEchart.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEcheckpoints.cpp
The-Chinese-U…/.../CSC3050-Project-2-simulator
ARCHIVEcheckpoints.h
The-Chinese-U…/.../CSC3050-Project-2-simulator
ARCHIVECHM1001 - Assignment 1 Problem.pdf
The-Chinese-Uni…/.../CHM1001-General-Chemistry
ARCHIVECHM1001 - Assignment 1.pdf
The-Chinese-Uni…/.../CHM1001-General-Chemistry
ARCHIVECHM1001 - Assignment 2 Problem.pdf
The-Chinese-Uni…/.../CHM1001-General-Chemistry
ARCHIVECHM1001 - Assignment 2.pdf
The-Chinese-Uni…/.../CHM1001-General-Chemistry
ARCHIVECHM1001 - Assignment 3 Problem.pdf
The-Chinese-Uni…/.../CHM1001-General-Chemistry
ARCHIVECHM1001 - Assignment 3.pdf
The-Chinese-Uni…/.../CHM1001-General-Chemistry
ARCHIVECHM1001 - Assignment 4 Problem.pdf
The-Chinese-Uni…/.../CHM1001-General-Chemistry
ARCHIVECHM1001 - Assignment 4.pdf
The-Chinese-Uni…/.../CHM1001-General-Chemistry
ARCHIVECHM1001 - Assignment 5 Problem.pdf
The-Chinese-Uni…/.../CHM1001-General-Chemistry
ARCHIVECHM1001 - Assignment 5.pdf
The-Chinese-Uni…/.../CHM1001-General-Chemistry
ARCHIVECHM1001 - Assignment 6 Problem.pdf
The-Chinese-Uni…/.../CHM1001-General-Chemistry
ARCHIVECHM1001 - Assignment 6.pdf
The-Chinese-Uni…/.../CHM1001-General-Chemistry
ARCHIVECHM1001.grades
The-Chinese-Uni…/.../CHM1001-General-Chemistry
ARCHIVEclassification_iris_testing_result_trial_1.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_testing_result_trial_10.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_testing_result_trial_2.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_testing_result_trial_3.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_testing_result_trial_4.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_testing_result_trial_5.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_testing_result_trial_6.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_testing_result_trial_7.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_testing_result_trial_8.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_testing_result_trial_9.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_training_result_trial_1.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_training_result_trial_10.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_training_result_trial_2.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_training_result_trial_3.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_training_result_trial_4.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_training_result_trial_5.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_training_result_trial_6.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_training_result_trial_7.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_training_result_trial_8.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEclassification_iris_training_result_trial_9.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVECMakeLists.txt
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVECMakeLists.txt
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVECMakeLists.txt
The-Chinese-University-of-Hong…/.../bucketsort
ARCHIVECMakeLists.txt
The-Chinese-University-of-Hong-…/.../mergesort
ARCHIVECMakeLists.txt
The-Chinese-University-of-H…/.../odd-even-sort
ARCHIVECMakeLists.txt
The-Chinese-University-of-Hong-…/.../quicksort
ARCHIVECMakeLists.txt
The-Chinese-University-of-Hong-Kong-S…/.../cpu
ARCHIVECMakeLists.txt
The-Chinese-University-of-Hong-Kong-S…/.../gpu
ARCHIVECMakeLists.txt
The-Chinese-University-of-Hong-Kong-S…/.../gpu
ARCHIVECMakeLists.txt
The-Chinese-U…/.../CSC3050-Project-1-assembler
ARCHIVECMakeLists.txt
The-Chinese-U…/.../CSC3050-Project-2-simulator
ARCHIVECMakeLists.txt
The-Chinese-University-…/.../CSC4005-Project-3
ARCHIVECMakeLists.txt
The-Chinese-University-of-Hong-K…/.../project1
ARCHIVECMakeLists.txt
The-Chinese-University-of-Hong-K…/.../project2
ARCHIVECMakeLists.txt
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVECMakeLists.txt.user
The-Chinese-U…/.../CSC3050-Project-1-assembler
ARCHIVECMakeLists.txt.user
The-Chinese-U…/.../CSC3050-Project-2-simulator
ARCHIVEcnn_classifier_openacc.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEcnn_classifier.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVECodeChef - BROKPHON.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - CHEFMATH.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - CHOPRT.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - EGRCAKE.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - FLOW001.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - FLOW002.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - FLOW004.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - FLOW010.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - GCDOPS.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - HMAPPY.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - JOHNY.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - MAXREM.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - MISSP.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - MSTICK.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - REDONE.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - SEAD.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - START01.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - STONES.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - TALCA.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - TSORT.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeChef - VK18.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1004D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1023B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1070F.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1083E with CHT.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1083E with LCT.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 109A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1101A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1109A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1113A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1131C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1131E.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1136B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1138A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1141C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1165A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1165E.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1166B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1167E.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 116A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 118A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 118D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1207F.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 121A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1270G.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1283D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 12B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1312E.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 135A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 137C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1387B1.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1401A.py
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1419D1.py
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1451A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 14C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1506D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 152C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 166E.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 182D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 189A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1997A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1997B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1997C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1997D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 1A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeforces - 2026A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeforces - 2026B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeforces - 2026C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeforces - 2026D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 208D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 20C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 225C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 233B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 235A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 242E.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 25D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 265B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 268C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 276D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 282A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 300C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 321C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 329D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 339D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 340A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 340D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 343B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 351E.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 363B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 363C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 364A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 367E.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 374C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 375B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 375D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 377A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 381A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 382C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 383E.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 385A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 401C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 407B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 414B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 427C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 429B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 433A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 455A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 459B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 459D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 463C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 466C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 466D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 474D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 475D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 476A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 478A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 478C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 486C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 489C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 489C.py
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 493D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 4A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 4C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 507B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 50A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 510A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 514D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 519E.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 52C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 538B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 538B.py
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 53C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 540B.py
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 545D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 557C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 557D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 559C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 567D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 570A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 580A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 584D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 588B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 597C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 599A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 620E.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 651B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 659D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 670B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 670D1.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 670D2.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 675E.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 680C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 682B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 686B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 69A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 700A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 706B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 712C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 713D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 735D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 749A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 750A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 754A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 762A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 764B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 768A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 768B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 779B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 794B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 798A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 804B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 832A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 842B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 844B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 872B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 876A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 877E.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 888B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 912A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 913C.py
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 932A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 96A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 980A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 987B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 995A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECodeForces - 9A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEcodeforceshuiwen.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEcollected-data.xlsx
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVEcollections.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVECombinatorics.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-1-src
ARCHIVECombinatorics.h
The-Chinese-Univ…/.../CSC3002-Assignment-1-src
ARCHIVEcomparison-image-filter.png
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVEcomparison-rgb-to-gray.png
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVEcomplex.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEconsole.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEconsolestreambuf.h
The-Chinese-University-of-Hong-Ko…/.../private
ARCHIVEconsoletext.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVECPU.v
The-Chinese-Univers…/.../CSC3050-Project-4-CPU
ARCHIVECSC1001 - Assignment 1 - q1.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 1 - q2.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 1 - q3.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 1 - q4.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 1 - q5.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 1 - q6.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 1 - report.pdf
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 1 Problem.pdf
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 2 - q1.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 2 - q2.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 2 - q3.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 2 - q4.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 2 - q5.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 2 - q6.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 2 - report.pdf
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 2 Problem.pdf
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 3 - q1.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 3 - q2.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 3 - q3.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 3 - report.pdf
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 3 Problem.pdf
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 4 - q1.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 4 - q2.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 4 - q3.py
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 4 - report.pdf
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001 - Assignment 4 Problem.pdf
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1001.grades
The…/.../CSC1001-Introduction-to-Computer-Science-Programming-Methodology
ARCHIVECSC1002 - Assignment 1 - report.pdf
The-Chin…/.../CSC1002-Computational-Laboratory
ARCHIVECSC1002 - Assignment 1 - source.py
The-Chin…/.../CSC1002-Computational-Laboratory
ARCHIVECSC1002 - Assignment 1 Problem.pdf
The-Chin…/.../CSC1002-Computational-Laboratory
ARCHIVECSC1002 - Assignment 2 - report.pdf
The-Chin…/.../CSC1002-Computational-Laboratory
ARCHIVECSC1002 - Assignment 2 - source.py
The-Chin…/.../CSC1002-Computational-Laboratory
ARCHIVECSC1002 - Assignment 2 Problem.pdf
The-Chin…/.../CSC1002-Computational-Laboratory
ARCHIVECSC1002.grades
The-Chin…/.../CSC1002-Computational-Laboratory
ARCHIVECSC3001 - Assignment 1 Problem.pdf
The-Chinese-…/.../CSC3001-Discrete-Mathematics
ARCHIVECSC3001 - Assignment 1.pdf
The-Chinese-…/.../CSC3001-Discrete-Mathematics
ARCHIVECSC3001 - Assignment 2 Problem.pdf
The-Chinese-…/.../CSC3001-Discrete-Mathematics
ARCHIVECSC3001 - Assignment 2.pdf
The-Chinese-…/.../CSC3001-Discrete-Mathematics
ARCHIVECSC3001 - Assignment 3 Problem.pdf
The-Chinese-…/.../CSC3001-Discrete-Mathematics
ARCHIVECSC3001 - Assignment 3.pdf
The-Chinese-…/.../CSC3001-Discrete-Mathematics
ARCHIVECSC3001 - Assignment 4 Problem.pdf
The-Chinese-…/.../CSC3001-Discrete-Mathematics
ARCHIVECSC3001 - Assignment 4.pdf
The-Chinese-…/.../CSC3001-Discrete-Mathematics
ARCHIVECSC3001 - Assignment 5 Problem.pdf
The-Chinese-…/.../CSC3001-Discrete-Mathematics
ARCHIVECSC3001 - Assignment 5.pdf
The-Chinese-…/.../CSC3001-Discrete-Mathematics
ARCHIVECSC3001.grades
The-Chinese-…/.../CSC3001-Discrete-Mathematics
ARCHIVECSC3002 - Assignment 1.pdf
The…/.../CSC3002-Introduction-to-Computer-Science-Programming-Paradigms
ARCHIVECSC3002 - Assignment 2.pdf
The…/.../CSC3002-Introduction-to-Computer-Science-Programming-Paradigms
ARCHIVECSC3002 - Assignment 3 Problem.pdf
The…/.../CSC3002-Introduction-to-Computer-Science-Programming-Paradigms
ARCHIVECSC3002 - Assignment 4 Problem.pdf
The…/.../CSC3002-Introduction-to-Computer-Science-Programming-Paradigms
ARCHIVECSC3002 - Assignment 5 Problem.pdf
The…/.../CSC3002-Introduction-to-Computer-Science-Programming-Paradigms
ARCHIVECSC3002 - Assignment 6 Problem.pdf
The…/.../CSC3002-Introduction-to-Computer-Science-Programming-Paradigms
ARCHIVECSC3002.grades
The…/.../CSC3002-Introduction-to-Computer-Science-Programming-Paradigms
ARCHIVECSC3050 - Project 1.pdf
The-Chinese…/.../CSC3050-Computer-Architecture
ARCHIVECSC3050 - Project 2.pdf
The-Chinese…/.../CSC3050-Computer-Architecture
ARCHIVECSC3050 - Project 3.pdf
The-Chinese…/.../CSC3050-Computer-Architecture
ARCHIVECSC3050 - Project 4.pdf
The-Chinese…/.../CSC3050-Computer-Architecture
ARCHIVECSC3050.grades
The-Chinese…/.../CSC3050-Computer-Architecture
ARCHIVECSC3100 - Assignment 1 Problem.pdf
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3100 - Assignment 1.pdf
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3100 - Assignment 2 - p1.cpp
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3100 - Assignment 2 - p2.cpp
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3100 - Assignment 2 - p3.cpp
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3100 - Assignment 2 - p4.cpp
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3100 - Assignment 2 - report.pdf
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3100 - Assignment 2 Problem.pdf
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3100 - Assignment 3 Problem.pdf
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3100 - Assignment 3.pdf
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3100 - Assignment 4 - p1.cpp
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3100 - Assignment 4 - p2.cpp
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3100 - Assignment 4 - p3.cpp
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3100 - Assignment 4 - p4.cpp
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3100 - Assignment 4 - report.pdf
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3100 - Assignment 4 Problem.pdf
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3100.grades
The-Chinese-Unive…/.../CSC3100-Data-Structures
ARCHIVECSC3150 - Assignment 1 Problem.pdf
The-Chinese-Uni…/.../CSC3150-Operating-Systems
ARCHIVECSC3150 - Assignment 2 Problem.pdf
The-Chinese-Uni…/.../CSC3150-Operating-Systems
ARCHIVECSC3150 - Assignment 3 Problem.pdf
The-Chinese-Uni…/.../CSC3150-Operating-Systems
ARCHIVECSC3150 - Assignment 4 Problem.pdf
The-Chinese-Uni…/.../CSC3150-Operating-Systems
ARCHIVECSC3150.grades
The-Chinese-Uni…/.../CSC3150-Operating-Systems
ARCHIVECSC3170 - Assignment 1 Problem.pdf
The-Chinese-Unive…/.../CSC3170-Database-System
ARCHIVECSC3170 - Assignment 1.pdf
The-Chinese-Unive…/.../CSC3170-Database-System
ARCHIVECSC3170 - Assignment 2 Problem.pdf
The-Chinese-Universi…/.../CSC3170-Assignment-2
ARCHIVECSC3170 - Assignment 3 Problem.pdf
The-Chinese-Unive…/.../CSC3170-Database-System
ARCHIVECSC3170 - Assignment 3.pdf
The-Chinese-Unive…/.../CSC3170-Database-System
ARCHIVECSC3170 - Assignment 4 Problem.pdf
The-Chinese-Unive…/.../CSC3170-Database-System
ARCHIVECSC3170 - Assignment 4.pdf
The-Chinese-Unive…/.../CSC3170-Database-System
ARCHIVECSC3170.grades
The-Chinese-Unive…/.../CSC3170-Database-System
ARCHIVECSC4001 - Assignment 1 Problem.pdf
The-Chinese-…/.../CSC4001-Software-Engineering
ARCHIVECSC4001 - Assignment 1.pdf
The-Chinese-…/.../CSC4001-Software-Engineering
ARCHIVECSC4001 - Assignment 2 Problem.pdf
The-Chinese-…/.../CSC4001-Software-Engineering
ARCHIVECSC4001 - Assignment 2.pdf
The-Chinese-…/.../CSC4001-Software-Engineering
ARCHIVECSC4001 - Assignment 3 Problem.pdf
The-Chinese-…/.../CSC4001-Software-Engineering
ARCHIVECSC4001 - Assignment 3.pdf
The-Chinese-…/.../CSC4001-Software-Engineering
ARCHIVECSC4001-Project-1-AST.pdf
The-Chinese-University-…/.../CSC4001-Project-1
ARCHIVECSC4001.grades
The-Chinese-…/.../CSC4001-Software-Engineering
ARCHIVECSC4005 - Project 1 Instruction.pdf
The-Chinese-…/.../CSC4005-Parallel-Programming
ARCHIVECSC4005 - Project 2 Instruction.pdf
The-Chinese-…/.../CSC4005-Parallel-Programming
ARCHIVECSC4005 - Project 3 Instruction.pdf
The-Chinese-…/.../CSC4005-Parallel-Programming
ARCHIVECSC4005 - Project 4 Instruction.pdf
The-Chinese-…/.../CSC4005-Parallel-Programming
ARCHIVECSC4005.grades
The-Chinese-…/.../CSC4005-Parallel-Programming
ARCHIVECSC4008 - Assignment 1 - Q1.ipynb
The-Chinese-Universi…/.../CSC4008-Assignment-1
ARCHIVECSC4008 - Assignment 1 - Q2.ipynb
The-Chinese-Universi…/.../CSC4008-Assignment-1
ARCHIVECSC4008 - Assignment 1 Problem.pdf
The-Ch…/.../CSC4008-Techniques-for-Data-Mining
ARCHIVECSC4008 - Assignment 1.pdf
The-Chinese-Universi…/.../CSC4008-Assignment-1
ARCHIVECSC4008 - Assignment 2 Problem.pdf
The-Ch…/.../CSC4008-Techniques-for-Data-Mining
ARCHIVECSC4008 - Assignment 2.pdf
The-Chinese-Universi…/.../CSC4008-Assignment-2
ARCHIVECSC4008 - Assignment 2.py
The-Chinese-Universi…/.../CSC4008-Assignment-2
ARCHIVECSC4008 - Assignment 3 - 1a.ipynb
The-Chinese-Universi…/.../CSC4008-Assignment-3
ARCHIVECSC4008 - Assignment 3 - 1b.ipynb
The-Chinese-Universi…/.../CSC4008-Assignment-3
ARCHIVECSC4008 - Assignment 3 Problem.pdf
The-Ch…/.../CSC4008-Techniques-for-Data-Mining
ARCHIVECSC4008 - Assignment 3.pdf
The-Chinese-Universi…/.../CSC4008-Assignment-3
ARCHIVECSC4008 - Assignment 4 Problem.pdf
The-Ch…/.../CSC4008-Techniques-for-Data-Mining
ARCHIVECSC4008 - Assignment 4.ipynb
The-Chinese-Universi…/.../CSC4008-Assignment-4
ARCHIVECSC4008 - Assignment 4.pdf
The-Chinese-Universi…/.../CSC4008-Assignment-4
ARCHIVECSC4008.grades
The-Ch…/.../CSC4008-Techniques-for-Data-Mining
ARCHIVECSC4120 - Written Homework 1.pdf
The…/.../CSC4120-Design-and-Analysis-of-Algorithms
ARCHIVECSC4120 - Written Homework 10.pdf
The…/.../CSC4120-Design-and-Analysis-of-Algorithms
ARCHIVECSC4120 - Written Homework 11.pdf
The…/.../CSC4120-Design-and-Analysis-of-Algorithms
ARCHIVECSC4120 - Written Homework 2.pdf
The…/.../CSC4120-Design-and-Analysis-of-Algorithms
ARCHIVECSC4120 - Written Homework 3.pdf
The…/.../CSC4120-Design-and-Analysis-of-Algorithms
ARCHIVECSC4120 - Written Homework 4.pdf
The…/.../CSC4120-Design-and-Analysis-of-Algorithms
ARCHIVECSC4120 - Written Homework 5.pdf
The…/.../CSC4120-Design-and-Analysis-of-Algorithms
ARCHIVECSC4120 - Written Homework 6.pdf
The…/.../CSC4120-Design-and-Analysis-of-Algorithms
ARCHIVECSC4120 - Written Homework 7.pdf
The…/.../CSC4120-Design-and-Analysis-of-Algorithms
ARCHIVECSC4120 - Written Homework 8.pdf
The…/.../CSC4120-Design-and-Analysis-of-Algorithms
ARCHIVECSC4120 - Written Homework 9.pdf
The…/.../CSC4120-Design-and-Analysis-of-Algorithms
ARCHIVECSC4120.grades
The…/.../CSC4120-Design-and-Analysis-of-Algorithms
ARCHIVECSC4130 - Assignment 1 Problem.pdf
The…/.../CSC4130-Introduction-to-Human-Computer-Interaction
ARCHIVECSC4130 - Assignment 2 Problem.pdf
The…/.../CSC4130-Introduction-to-Human-Computer-Interaction
ARCHIVECSC4130 - Assignment 3 Problem.pdf
The…/.../CSC4130-Introduction-to-Human-Computer-Interaction
ARCHIVECSC4130 - Assignment 4 Problem.pdf
The…/.../CSC4130-Introduction-to-Human-Computer-Interaction
ARCHIVECSC4130.grades
The…/.../CSC4130-Introduction-to-Human-Computer-Interaction
ARCHIVECSES - 1096 Throwing Dice.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECSES - 1628 Meet in the Middle.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECSES - 1702 Tree Traversals.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECSES - 1722 Fibonacci Numbers.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECSES - 1726 Moving Robots.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECSES - 1729 Stick Game.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECSES - 1730 Nim Game I.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECSES - 2136 Hamming Distance.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECSES - 2137 Beautiful Subgrids.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVECSES - 2138 Reachable Nodes.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEcses1622.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEcses1623.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEcses1624.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEcuda_PartA.cu
The-Chinese-University-of-Hong-Kong-S…/.../gpu
ARCHIVEcuda_PartB.cu
The-Chinese-University-of-Hong-Kong-S…/.../gpu
ARCHIVEcuda.cu
The-Chinese-University-of-Hong-Kong-S…/.../gpu
ARCHIVEd.cpp
The-Chinese-Universi…/.../DDA6050-Assignment-2
ARCHIVEd.cpp
The-Chinese-Universi…/.../DDA6050-Assignment-4
ARCHIVEd3-timer.v1.min.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEd3.v5.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEd3.v6.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEd3.v6.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEd3.v6.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEd3.v6.min.js
The-Chinese-University-of-Hong-Kong-Sh…/.../js
ARCHIVEd3.v6.min.js
The-Chinese-University-of-Hong-Kong-Sh…/.../js
ARCHIVEdata_bank_data.csv
The-Chinese-University-of-Hong-Kong-…/.../data
ARCHIVEdata.bin
The-Chinese-University-of-Hong-Kong…/.../bonus
ARCHIVEdata.bin
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVEdata.bin
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVEdata.txt
The-Chinese-University-of-Hong-Ko…/.../data-Q1
ARCHIVEdawglexicon.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEdb_insert.sql
The-Chinese-Universi…/.../CSC3170-Assignment-2
ARCHIVEDDA2003 - Assignment 1.pdf
The-Chinese-Univ…/.../DDA2003-Visual-Analytics
ARCHIVEDDA2003 - Assignment 2.pdf
The-Chinese-Univ…/.../DDA2003-Visual-Analytics
ARCHIVEDDA2003 - Assignment 3.pdf
The-Chinese-Univ…/.../DDA2003-Visual-Analytics
ARCHIVEDDA2003 - Assignment 4.pdf
The-Chinese-Univ…/.../DDA2003-Visual-Analytics
ARCHIVEDDA2003 - Assignment 5.pdf
The-Chinese-Univ…/.../DDA2003-Visual-Analytics
ARCHIVEDDA2003.grades
The-Chinese-Univ…/.../DDA2003-Visual-Analytics
ARCHIVEDDA3020 - Assignment 1 - q2-1.py
The-Chinese-Universi…/.../DDA3020-Assignment-1
ARCHIVEDDA3020 - Assignment 1 - q2-2.py
The-Chinese-Universi…/.../DDA3020-Assignment-1
ARCHIVEDDA3020 - Assignment 1 Problem.pdf
The-Chinese-Univ…/.../DDA3020-Machine-Learning
ARCHIVEDDA3020 - Assignment 1.pdf
The-Chinese-Universi…/.../DDA3020-Assignment-1
ARCHIVEDDA3020 - Assignment 2 Problem.pdf
The-Chinese-Univ…/.../DDA3020-Machine-Learning
ARCHIVEDDA3020 - Assignment 2.ipynb
The-Chinese-Universi…/.../DDA3020-Assignment-2
ARCHIVEDDA3020 - Assignment 2.pdf
The-Chinese-Universi…/.../DDA3020-Assignment-2
ARCHIVEDDA3020 - Assignment 3 Problem.pdf
The-Chinese-Univ…/.../DDA3020-Machine-Learning
ARCHIVEDDA3020 - Assignment 3.ipynb
The-Chinese-Universi…/.../DDA3020-Assignment-3
ARCHIVEDDA3020 - Assignment 3.pdf
The-Chinese-Universi…/.../DDA3020-Assignment-3
ARCHIVEDDA3020 - Assignment 4 Problem.pdf
The-Chinese-Univ…/.../DDA3020-Machine-Learning
ARCHIVEDDA3020 - Assignment 4.ipynb
The-Chinese-Universi…/.../DDA3020-Assignment-4
ARCHIVEDDA3020 - Assignment 4.pdf
The-Chinese-Universi…/.../DDA3020-Assignment-4
ARCHIVEDDA3020.grades
The-Chinese-Univ…/.../DDA3020-Machine-Learning
ARCHIVEDDA4210 - Assignment 1.pdf
The-Chi…/.../DDA4210-Advanced-Machine-Learning
ARCHIVEDDA4210 - Assignment 2.pdf
The-Chi…/.../DDA4210-Advanced-Machine-Learning
ARCHIVEDDA4210 - Assignment 3.pdf
The-Chi…/.../DDA4210-Advanced-Machine-Learning
ARCHIVEDDA4210.grades
The-Chi…/.../DDA4210-Advanced-Machine-Learning
ARCHIVEDDA6050 - Assignment 1 Problem.pdf
The-Chines…/.../DDA6050-Analysis-of-Algorithms
ARCHIVEDDA6050 - Assignment 1.pdf
The-Chines…/.../DDA6050-Analysis-of-Algorithms
ARCHIVEDDA6050 - Assignment 2 Problem.pdf
The-Chines…/.../DDA6050-Analysis-of-Algorithms
ARCHIVEDDA6050 - Assignment 3 Problem.pdf
The-Chines…/.../DDA6050-Analysis-of-Algorithms
ARCHIVEDDA6050 - Assignment 3.pdf
The-Chines…/.../DDA6050-Analysis-of-Algorithms
ARCHIVEDDA6050 - Assignment 4 Problem.pdf
The-Chines…/.../DDA6050-Analysis-of-Algorithms
ARCHIVEDDA6050.grades
The-Chines…/.../DDA6050-Analysis-of-Algorithms
ARCHIVEdda6050a2q1.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEdda6050a2q12023.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEdda6050a2q2.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEdda6050a2q22023.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEdda6050a2q3.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEdda6050a2q32023.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEdda6050a2q4.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEdda6050a2q42023.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEdda6050a2q5.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEdda6050a4q12023fall.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEdda6050a4q2.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEdda6050a4q22023fall.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEdda6050a4q3.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEdda6050a4q32023fall.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEdda6050a4q4.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEdda6050a4q42023fall.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEDDA6205 - Assignment 1.pdf
The…/.../DDA6205-Game-Theory-and-Its-Applications
ARCHIVEDDA6205 - Assignment 2.pdf
The…/.../DDA6205-Game-Theory-and-Its-Applications
ARCHIVEDDA6205 - Assignment 3.pdf
The…/.../DDA6205-Game-Theory-and-Its-Applications
ARCHIVEDDA6205.grades
The…/.../DDA6205-Game-Theory-and-Its-Applications
ARCHIVEdeque.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEdesc.txt
Project-Euler/.../1
ARCHIVEdesc.txt
Project-Euler/.../10
ARCHIVEdesc.txt
Project-Euler/.../11
ARCHIVEdesc.txt
Project-Euler/.../12
ARCHIVEdesc.txt
Project-Euler/.../13
ARCHIVEdesc.txt
Project-Euler/.../14
ARCHIVEdesc.txt
Project-Euler/.../15
ARCHIVEdesc.txt
Project-Euler/.../16
ARCHIVEdesc.txt
Project-Euler/.../17
ARCHIVEdesc.txt
Project-Euler/.../18
ARCHIVEdesc.txt
Project-Euler/.../19
ARCHIVEdesc.txt
Project-Euler/.../2
ARCHIVEdesc.txt
Project-Euler/.../20
ARCHIVEdesc.txt
Project-Euler/.../21
ARCHIVEdesc.txt
Project-Euler/.../22
ARCHIVEdesc.txt
Project-Euler/.../23
ARCHIVEdesc.txt
Project-Euler/.../24
ARCHIVEdesc.txt
Project-Euler/.../25
ARCHIVEdesc.txt
Project-Euler/.../26
ARCHIVEdesc.txt
Project-Euler/.../27
ARCHIVEdesc.txt
Project-Euler/.../28
ARCHIVEdesc.txt
Project-Euler/.../29
ARCHIVEdesc.txt
Project-Euler/.../3
ARCHIVEdesc.txt
Project-Euler/.../30
ARCHIVEdesc.txt
Project-Euler/.../31
ARCHIVEdesc.txt
Project-Euler/.../32
ARCHIVEdesc.txt
Project-Euler/.../33
ARCHIVEdesc.txt
Project-Euler/.../34
ARCHIVEdesc.txt
Project-Euler/.../35
ARCHIVEdesc.txt
Project-Euler/.../36
ARCHIVEdesc.txt
Project-Euler/.../37
ARCHIVEdesc.txt
Project-Euler/.../4
ARCHIVEdesc.txt
Project-Euler/.../5
ARCHIVEdesc.txt
Project-Euler/.../6
ARCHIVEdesc.txt
Project-Euler/.../7
ARCHIVEdesc.txt
Project-Euler/.../8
ARCHIVEdesc.txt
Project-Euler/.../9
ARCHIVEdiff.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEdirection.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEdiscriminatorModel.py
The…/.../text-based-image-generation-for-cuhksz-buildings
ARCHIVEdraw-map.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEechoinputstreambuf.h
The-Chinese-University-of-Hong-Ko…/.../private
ARCHIVEeducation.json
The-Chinese-University-of-Hong-Kong-…/.../data
ARCHIVEego-facebook.txt
The-Chinese-University-of-Hong-Ko…/.../data-Q2
ARCHIVEEIE2050 - Homework 1 - base_convertion.py
The-Ch…/.../EIE2050-Digital-Logics-and-Systems
ARCHIVEEIE2050 - Homework 1.pdf
The-Ch…/.../EIE2050-Digital-Logics-and-Systems
ARCHIVEEIE2050 - Homework 2.pdf
The-Ch…/.../EIE2050-Digital-Logics-and-Systems
ARCHIVEEIE2050 - Homework 3.pdf
The-Ch…/.../EIE2050-Digital-Logics-and-Systems
ARCHIVEEIE2050 - Homework 4.pdf
The-Ch…/.../EIE2050-Digital-Logics-and-Systems
ARCHIVEEIE2050 - Homework 5.pdf
The-Ch…/.../EIE2050-Digital-Logics-and-Systems
ARCHIVEEIE2050 - Homework 6.pdf
The-Ch…/.../EIE2050-Digital-Logics-and-Systems
ARCHIVEEIE2050 - Homework 7.pdf
The-Ch…/.../EIE2050-Digital-Logics-and-Systems
ARCHIVEEIE2050 - Homework Problems.pdf
The-Ch…/.../EIE2050-Digital-Logics-and-Systems
ARCHIVEEIE2050.grades
The-Ch…/.../EIE2050-Digital-Logics-and-Systems
ARCHIVEerror-loss-results.png
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVEerror-loss-results.xlsx
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVEerror.h
The-Chinese-University-of-Hong-Kon…/.../system
ARCHIVEexceptions.h
The-Chinese-University-of-Hong-Kon…/.../system
ARCHIVEexecution-time-results.png
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVEexecution-time-results.xlsx
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVEfile_system.cu
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVEfile_system.h
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVEfilelib.h
The-Chinese-University-of-Hong-Kong-Sh…/.../io
ARCHIVEFindDNAMatch.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-1-src
ARCHIVEFindDNAMatch.h
The-Chinese-Univ…/.../CSC3002-Assignment-1-src
ARCHIVEfloating.c
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVEforceDirectedGraph.js
The-Chinese-University-of-Hong-Kong-Sh…/.../js
ARCHIVEforeach.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEforeachpatch.h
The-Chinese-University-of-Hong-Ko…/.../private
ARCHIVEforwardingstreambuf.h
The-Chinese-University-of-Hong-Ko…/.../private
ARCHIVEfunctional.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEgbrowserpane.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgbufferedimage.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgbutton.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgcanvas.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgcheckbox.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgchooser.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgclipboard.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgcolor.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgcolorchooser.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgconsolewindow.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgcontainer.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgdiffgui.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgdiffimage.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgdownloader.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgdrawingsurface.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgeneratorModel.py
The…/.../text-based-image-generation-for-cuhksz-buildings
ARCHIVEgevent.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgeventqueue.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgevents.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgfilechooser.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgfont.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgfontchooser.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgformattedpane.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEginputpanel.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEginteractor.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEginteractors.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEglabel.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEglayout.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgmath.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEgobjects.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgobservable.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgoptionpane.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgradiobutton.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgrammy.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEgraph-full.txt
The-Chinese-University-of-Hong-Kong-…/.../data
ARCHIVEgraph-small.txt
The-Chinese-University-of-Hong-Kong-…/.../data
ARCHIVEgraph.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEgraphtypes.h
The-Chinese-Univ…/.../CSC3002-Assignment-6-src
ARCHIVEgrid.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEgridlocation.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEgscrollbar.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgscrollpane.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgslider.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgspacer.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgtable.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgtextarea.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgtextfield.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgthread.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgtimer.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgtypes.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEgwindow.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEGym - 100185A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEGym - 100185B.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEGym - 100733A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEGym - 100733C.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEGym - 100733D.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEGym - 100733E.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEGym - 100886G.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEGym - 101021A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEGym - 101401G.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEGym - 101804G.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEGym - 102411A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEGym - 103536A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEGymnasticsJudge.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-3-src
ARCHIVEGymnasticsJudge.h
The-Chinese-Univ…/.../CSC3002-Assignment-3-src
ARCHIVEHackerRank - dreamplay-and-clubbing.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEHackerRank - find-the-running-median.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEHackerRank - handshake.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEHackerRank - maximum-draws.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEhangup.c
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVEhashcode.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEhashmap.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEhashset.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEhdu1253.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEhdu1599.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEheadless.h
The-Chinese-University-of-Hong-Ko…/.../private
ARCHIVEHFractal.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-4-src
ARCHIVEHFractal.h
The-Chinese-Univ…/.../CSC3002-Assignment-4-src
ARCHIVEhttpserver
The-Chinese-University-of-Hon…/.../thread_pool
ARCHIVEhttpserver.c
The-Chinese-University-of-Hon…/.../thread_pool
ARCHIVEhttpserver.o
The-Chinese-University-of-Hon…/.../thread_pool
ARCHIVEhw2.cpp
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVEillegal_instr.c
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVEimages-tree-structure.txt
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVEindex.css
The-Chinese-Universi…/.../CSC4130-Assignment-1
ARCHIVEindex.css
The-Chinese-Universi…/.../CSC4130-Assignment-2
ARCHIVEindex.css
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEindex.html
The-Chinese-Universi…/.../CSC4130-Assignment-1
ARCHIVEindex.html
The-Chinese-Universi…/.../CSC4130-Assignment-2
ARCHIVEindex.html
The-Chinese-Universi…/.../CSC4130-Assignment-3
ARCHIVEindex.html
The-Chinese-Universi…/.../DDA2003-Assignment-1
ARCHIVEindex.html
The-Chinese-Universi…/.../DDA2003-Assignment-2
ARCHIVEindex.html
The-Chinese-Universi…/.../DDA2003-Assignment-3
ARCHIVEindex.html
The-Chinese-Universi…/.../DDA2003-Assignment-4
ARCHIVEindex.html
The-Chinese-Universi…/.../DDA2003-Assignment-5
ARCHIVEindex.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEindex.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEindex.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEinit.h
The-Chinese-University-of-Hong-Ko…/.../private
ARCHIVEinitstudent.h
The-Chinese-University-of-Hong-Ko…/.../private
ARCHIVEInstructionRAM.v
The-Chinese-Univers…/.../CSC3050-Project-4-CPU
ARCHIVEintarray.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-3-src
ARCHIVEintarray.h
The-Chinese-Univ…/.../CSC3002-Assignment-3-src
ARCHIVEinterrupt.c
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVEintrange.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEioi20081a.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEisosurface_rendering_with_outline.png
The-Chinese-Universi…/.../DDA2003-Assignment-6
ARCHIVEKattis - allpairspath.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - caveexploration.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - goingtoseed.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - gold.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - gottacatchemall.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - grandpabernie.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - growingupishardtodo.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - joinstrings.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - knightjump.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - modulodatastructures.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - polygonarea.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - quickbrownfox.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - shesallyak.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - shortestpath1.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - simplicity.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - sparklesseven.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - teque.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - thelastlaugh.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEKattis - welcomehard.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEkill.c
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVEKnightsTour.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-4-src
ARCHIVEKnightsTour.h
The-Chinese-Univ…/.../CSC3002-Assignment-4-src
ARCHIVElabeltable.cpp
The-Chinese-U…/.../CSC3050-Project-2-simulator
ARCHIVElabelTable.cpp
The-Chinese-U…/.../CSC3050-Project-1-assembler
ARCHIVElabeltable.h
The-Chinese-U…/.../CSC3050-Project-2-simulator
ARCHIVElabelTable.h
The-Chinese-U…/.../CSC3050-Project-1-assembler
ARCHIVELena-smooth-cuda.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-mpi-1.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-mpi-16.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-mpi-2.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-mpi-32.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-mpi-4.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-mpi-8.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-mpi-openmp-16x2.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-mpi-openmp-1x32.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-mpi-openmp-2x16.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-mpi-openmp-32x1.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-mpi-openmp-4x8.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-mpi-openmp-8x4.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-openacc.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-openmp-1.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-openmp-16.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-openmp-2.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-openmp-32.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-openmp-4.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-openmp-8.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-pthread-1.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-pthread-16.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-pthread-2.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-pthread-32.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-pthread-4.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-pthread-8.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-sequential.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth-simd.jpg
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVELena-smooth.png
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVElexicon.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVElibhttp.c
The-Chinese-University-of-Hon…/.../thread_pool
ARCHIVElibhttp.h
The-Chinese-University-of-Hon…/.../thread_pool
ARCHIVElibhttp.o
The-Chinese-University-of-Hon…/.../thread_pool
ARCHIVELightOJ - 1023.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVELightOJ - 1024.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVELightOJ - 1082.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVELightOJ - 1112.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVElimitoutputstreambuf.h
The-Chinese-University-of-Hong-Ko…/.../private
ARCHIVElinkedhashmap.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVElinkedhashset.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVElinkedlist.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVElocality.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVElogo.svg
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEloj10000.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEloj10005.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEloj10007.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEloj10050.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEloj10127.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEloj10175.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEloj10176.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEloj104.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEloj107.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEluogu1129.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEluogu1706.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEluogu2386.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEmain.cpp
The-Chinese-U…/.../CSC3050-Project-1-assembler
ARCHIVEmain.cpp
The-Chinese-U…/.../CSC3050-Project-2-simulator
ARCHIVEmain.cu
The-Chinese-University-of-Hong-Kong…/.../bonus
ARCHIVEmain.cu
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVEmain.cu
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVEmain.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEmain.js
The-Chinese-University-of-Hong-Kong-Sh…/.../js
ARCHIVEmain.js
The-Chinese-University-of-Hong-Kong-Sh…/.../js
ARCHIVEmain.py
Project-Euler/.../1
ARCHIVEmain.py
Project-Euler/.../10
ARCHIVEmain.py
Project-Euler/.../11
ARCHIVEmain.py
Project-Euler/.../12
ARCHIVEmain.py
Project-Euler/.../13
ARCHIVEmain.py
Project-Euler/.../14
ARCHIVEmain.py
Project-Euler/.../15
ARCHIVEmain.py
Project-Euler/.../16
ARCHIVEmain.py
Project-Euler/.../17
ARCHIVEmain.py
Project-Euler/.../18
ARCHIVEmain.py
Project-Euler/.../19
ARCHIVEmain.py
Project-Euler/.../2
ARCHIVEmain.py
Project-Euler/.../20
ARCHIVEmain.py
Project-Euler/.../21
ARCHIVEmain.py
Project-Euler/.../22
ARCHIVEmain.py
Project-Euler/.../23
ARCHIVEmain.py
Project-Euler/.../24
ARCHIVEmain.py
Project-Euler/.../25
ARCHIVEmain.py
Project-Euler/.../26
ARCHIVEmain.py
Project-Euler/.../27
ARCHIVEmain.py
Project-Euler/.../28
ARCHIVEmain.py
Project-Euler/.../29
ARCHIVEmain.py
Project-Euler/.../3
ARCHIVEmain.py
Project-Euler/.../30
ARCHIVEmain.py
Project-Euler/.../31
ARCHIVEmain.py
Project-Euler/.../32
ARCHIVEmain.py
Project-Euler/.../33
ARCHIVEmain.py
Project-Euler/.../34
ARCHIVEmain.py
Project-Euler/.../35
ARCHIVEmain.py
Project-Euler/.../36
ARCHIVEmain.py
Project-Euler/.../37
ARCHIVEmain.py
Project-Euler/.../4
ARCHIVEmain.py
Project-Euler/.../5
ARCHIVEmain.py
Project-Euler/.../6
ARCHIVEmain.py
Project-Euler/.../7
ARCHIVEmain.py
Project-Euler/.../8
ARCHIVEmain.py
Project-Euler/.../9
ARCHIVEmain.py
The-Chinese-University-of-…/.../party-together
ARCHIVEMainMemory.v
The-Chinese-Univers…/.../CSC3050-Project-4-CPU
ARCHIVEMake.txt
The-Chinese-Uni…/.../CSC3050-Project-3-verilog
ARCHIVEMake.txt
The-Chinese-Univers…/.../CSC3050-Project-4-CPU
ARCHIVEMakefile
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVEMakefile
The-Chinese-University-of-Hong-K…/.../program2
ARCHIVEMakefile
The-Chinese-University-of-Hon…/.../thread_pool
ARCHIVEmanaged.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEmap.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEMAT1001 - Homework Problems.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Homework Week 1.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Homework Week 10.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Homework Week 11.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Homework Week 12.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Homework Week 2.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Homework Week 3.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Homework Week 4.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Homework Week 5.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Homework Week 6.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Homework Week 7.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Homework Week 8.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Homework Week 9.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 1 Problem.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 1.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 10 Problem.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 10.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 11 Problem.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 11.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 12 Problem.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 12.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 2 Problem.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 2.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 3 Problem.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 3.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 4 Problem.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 4.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 5 Problem.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 5.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 6 Problem.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 6.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 7 Problem.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 7.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 8 Problem.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 8.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 9 Problem.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001 - Quiz 9.pdf
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1001.grades
The-Chinese-University…/.../MAT1001-Calculus-I
ARCHIVEMAT1002 - Homework Problems.pdf
The-Chinese-Universit…/.../MAT1002-Calculus-II
ARCHIVEMAT1002 - Homework Week 1.pdf
The-Chinese-Universit…/.../MAT1002-Calculus-II
ARCHIVEMAT1002 - Homework Week 10.pdf
The-Chinese-Universit…/.../MAT1002-Calculus-II
ARCHIVEMAT1002 - Homework Week 11.pdf
The-Chinese-Universit…/.../MAT1002-Calculus-II
ARCHIVEMAT1002 - Homework Week 12.pdf
The-Chinese-Universit…/.../MAT1002-Calculus-II
ARCHIVEMAT1002 - Homework Week 2.pdf
The-Chinese-Universit…/.../MAT1002-Calculus-II
ARCHIVEMAT1002 - Homework Week 3.pdf
The-Chinese-Universit…/.../MAT1002-Calculus-II
ARCHIVEMAT1002 - Homework Week 4.pdf
The-Chinese-Universit…/.../MAT1002-Calculus-II
ARCHIVEMAT1002 - Homework Week 5.pdf
The-Chinese-Universit…/.../MAT1002-Calculus-II
ARCHIVEMAT1002 - Homework Week 6.pdf
The-Chinese-Universit…/.../MAT1002-Calculus-II
ARCHIVEMAT1002 - Homework Week 7.pdf
The-Chinese-Universit…/.../MAT1002-Calculus-II
ARCHIVEMAT1002 - Homework Week 8.pdf
The-Chinese-Universit…/.../MAT1002-Calculus-II
ARCHIVEMAT1002 - Homework Week 9.pdf
The-Chinese-Universit…/.../MAT1002-Calculus-II
ARCHIVEMAT1002.grades
The-Chinese-Universit…/.../MAT1002-Calculus-II
ARCHIVEMAT2040 - Assignment 1 Problem.pdf
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT2040 - Assignment 1.pdf
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT2040 - Assignment 2 Problem.pdf
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT2040 - Assignment 2.pdf
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT2040 - Assignment 3 Problem.pdf
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT2040 - Assignment 3.pdf
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT2040 - Assignment 4 Problem.pdf
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT2040 - Assignment 4.pdf
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT2040 - Assignment 5 Problem.pdf
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT2040 - Assignment 5.pdf
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT2040 - Assignment 6 Problem.pdf
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT2040 - Assignment 6.pdf
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT2040 - Assignment 7 Problem.pdf
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT2040 - Assignment 7.pdf
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT2040 - Assignment 8 Problem.pdf
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT2040 - Assignment 8.pdf
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT2040.grades
The-Chinese-Univer…/.../MAT2040-Linear-Algebra
ARCHIVEMAT3007 - Assignment 1 Problem.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 1.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 2 Problem.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 2.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 3 Problem.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 3.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 4 Problem.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 4.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 5 Problem.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 5.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 6 Problem.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 6.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 7 Problem.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 7.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 8 Problem.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 9 - P1.py
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 9 - P2.ipynb
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 9 Problem.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007 - Assignment 9.pdf
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEMAT3007.grades
The-Chinese-Universi…/.../MAT3007-Optimization
ARCHIVEmatrices.png
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVEmatrix.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEmatrix.hpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEmatrix1.txt
The-Chinese-University-of-Hong-K…/.../matrices
ARCHIVEmatrix2.txt
The-Chinese-University-of-Hong-K…/.../matrices
ARCHIVEmatrix3.txt
The-Chinese-University-of-Hong-K…/.../matrices
ARCHIVEmatrix4.txt
The-Chinese-University-of-Hong-K…/.../matrices
ARCHIVEmatrix5.txt
The-Chinese-University-of-Hong-K…/.../matrices
ARCHIVEmatrix6.txt
The-Chinese-University-of-Hong-K…/.../matrices
ARCHIVEmatrix7.txt
The-Chinese-University-of-Hong-K…/.../matrices
ARCHIVEmatrix8.txt
The-Chinese-University-of-Hong-K…/.../matrices
ARCHIVEmemory.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEMergeSort.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-4-src
ARCHIVEMergeSort.h
The-Chinese-Univ…/.../CSC3002-Assignment-4-src
ARCHIVEmiserables.json
The-Chinese-University-of-Hong-Kong-…/.../data
ARCHIVEml_ext.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEml_ext.hpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEml_openacc.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEml_openacc.hpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEmorsecode.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-2-src
ARCHIVEmorsecode.h
The-Chinese-Univ…/.../CSC3002-Assignment-2-src
ARCHIVEmpi_openmp_PartB.cpp
The-Chinese-University-of-Hong-Kong-S…/.../cpu
ARCHIVEmpi_PartA.cpp
The-Chinese-University-of-Hong-Kong-S…/.../cpu
ARCHIVEmpi_PartB.cpp
The-Chinese-University-of-Hong-Kong-S…/.../cpu
ARCHIVEmpi.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEmpi.cpp
The-Chinese-University-of-Hong…/.../bucketsort
ARCHIVEmpi.cpp
The-Chinese-University-of-H…/.../odd-even-sort
ARCHIVEmpi.cpp
The-Chinese-University-of-Hong-…/.../quicksort
ARCHIVEmtsp_dp.py
The-Chinese-University-of-…/.../party-together
ARCHIVEmultimain.h
The-Chinese-University-of-Hong-Ko…/.../private
ARCHIVEmy_weather_data.json
The-Chinese-University-of-Hong-Kong-…/.../data
ARCHIVEnaive.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEnn_classifier_openacc.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEnn_classifier.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEnn_openacc.txt
The-Chinese-University-of-Hong-…/.../profiling
ARCHIVEnormal.c
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVEnote.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEobservable.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEopenacc_PartA.cpp
The-Chinese-University-of-Hong-Kong-S…/.../gpu
ARCHIVEopenacc_PartB.cpp
The-Chinese-University-of-Hong-Kong-S…/.../gpu
ARCHIVEopenmp_PartA.cpp
The-Chinese-University-of-Hong-Kong-S…/.../cpu
ARCHIVEopenmp_PartB.cpp
The-Chinese-University-of-Hong-Kong-S…/.../cpu
ARCHIVEopenmp.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEos.h
The-Chinese-University-of-Hong-Kon…/.../system
ARCHIVEP1PriorityQueue.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-6-src
ARCHIVEP1PriorityQueue.h
The-Chinese-Univ…/.../CSC3002-Assignment-6-src
ARCHIVEP2Traverse.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-6-src
ARCHIVEP2Traverse.h
The-Chinese-Univ…/.../CSC3002-Assignment-6-src
ARCHIVEparallel.cpp
The-Chinese-University-of-Hong-…/.../mergesort
ARCHIVEparallel.cpp
The-Chinese-University-of-Hong-…/.../quicksort
ARCHIVEperformance-evaluation-1024x1024.png
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVEperformance-evaluation-2048x2048.png
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVEphase1.cpp
The-Chinese-U…/.../CSC3050-Project-1-assembler
ARCHIVEphase1.cpp
The-Chinese-U…/.../CSC3050-Project-2-simulator
ARCHIVEphase1.h
The-Chinese-U…/.../CSC3050-Project-1-assembler
ARCHIVEphase1.h
The-Chinese-U…/.../CSC3050-Project-2-simulator
ARCHIVEphase2.cpp
The-Chinese-U…/.../CSC3050-Project-1-assembler
ARCHIVEphase2.cpp
The-Chinese-U…/.../CSC3050-Project-2-simulator
ARCHIVEphase2.h
The-Chinese-U…/.../CSC3050-Project-1-assembler
ARCHIVEphase2.h
The-Chinese-U…/.../CSC3050-Project-2-simulator
ARCHIVEPHY1001 - Assignment 1 Problem.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 1.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 10 Problem.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 10.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 11 Problem.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 11.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 12 Problem.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 12.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 13 Problem.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 13.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 14 Problem.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 14.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 15 Problem.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 15.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 2 Problem.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 2.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 3 Problem.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 3.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 4 Problem.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 4.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 5 Problem.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 5.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 6 Problem.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 6.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 7 Problem.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 7.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 8 Problem.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 8.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 9 Problem.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001 - Assignment 9.pdf
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1001.grades
The-Chinese-University-…/.../PHY1001-Mechanics
ARCHIVEPHY1002 - Error Analysis Homework Problem.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Error Analysis Homework.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Experiment 1 Report.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Experiment 1 Template - Conservation of Angular Momentum.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Experiment 2 Report.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Experiment 2 Template - Large Amplitude Pendulum.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Experiment 3 Report.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Experiment 3 Template - Conservation of Energy.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Experiment 4 Report.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Experiment 4 Template - Driven Damped Harmonic Oscillations.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Experiment 5 Report.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Experiment 5 Template - Rotational Inertia.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Experiment 6 Report.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Experiment 6 Template - Centripetal Force.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Experiment 7 Report.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Experiment 7 Template - Conservation of Momentum.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Experiment 8 Report.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002 - Experiment 8 Template - Resonance Air Column.pdf
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEPHY1002.grades
The-Chinese-Un…/.../PHY1002-Physics-Laboratory
ARCHIVEpipe.c
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVEplainconsole.h
The-Chinese-University-of-Hong-Kong-Sh…/.../io
ARCHIVEpoint.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEpqueue.h
The-Chinese-Univ…/.../CSC3002-Assignment-5-src
ARCHIVEpqueue.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEprecompiled.h
The-Chinese-University-of-Hong-Ko…/.../private
ARCHIVEpreprocessing.py
The…/.../text-based-image-generation-for-cuhksz-buildings
ARCHIVEpriorityqueue.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEprog_assignment_1.ipynb
The…/.../CSC4120-Design-and-Analysis-of-Algorithms
ARCHIVEprog_assignment_2.ipynb
The…/.../CSC4120-Design-and-Analysis-of-Algorithms
ARCHIVEprog_assignment_3.ipynb
The…/.../CSC4120-Design-and-Analysis-of-Algorithms
ARCHIVEprogram1.c
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVEprogram2.c
The-Chinese-University-of-Hong-K…/.../program2
ARCHIVEProject1-PartA-Results.txt
The-Chinese-University-of…/.../Gray (no image)
ARCHIVEProject1-PartB-Results.txt
The-Chinese-Univers…/.../20K-smooth (no image)
ARCHIVEProject1-PartB-Results.txt
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVEProject2-Results-Perf.txt
The-Chinese-University-of-Hong-K…/.../project2
ARCHIVEProject2-Results.txt
The-Chinese-University-of-Hong-K…/.../project2
ARCHIVEProject3-Perf-Result-task-1.txt
The-Chinese-University-of-Hong-…/.../profiling
ARCHIVEProject3-Perf-Result-task-2.txt
The-Chinese-University-of-Hong-…/.../profiling
ARCHIVEProject3-Perf-Result-task-3.txt
The-Chinese-University-of-Hong-…/.../profiling
ARCHIVEProject3-Perf-Result-task-4.txt
The-Chinese-University-of-Hong-…/.../profiling
ARCHIVEProject3-Perf-Result-task-5.txt
The-Chinese-University-of-Hong-…/.../profiling
ARCHIVEProject3-Result.txt
The-Chinese-University-…/.../CSC4005-Project-3
ARCHIVEpthp_from_tsp.py
The-Chinese-University-of-…/.../party-together
ARCHIVEpthread_PartA.cpp
The-Chinese-University-of-Hong-Kong-S…/.../cpu
ARCHIVEpthread_PartB.cpp
The-Chinese-University-of-Hong-Kong-S…/.../cpu
ARCHIVEptp_cli.py
The-Chinese-University-of-…/.../party-together
ARCHIVEptp_solver.py
The-Chinese-University-of-…/.../party-together
ARCHIVEpuzzle.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEQ1.sql
The-Chinese-Universi…/.../CSC3170-Assignment-2
ARCHIVEQ1.sql
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEQ10.sql
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEQ11.sql
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEQ12.sql
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEQ13.sql
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEQ14.sql
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEQ15.sql
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEQ16.sql
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEQ2.sql
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEQ3.sql
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEQ4.sql
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEQ5.sql
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEQ6.sql
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEQ7.sql
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEQ8.sql
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEQ9.sql
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEqtgui.h
The-Chinese-University-of-Hong-K…/.../graphics
ARCHIVEqueue.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEquit.c
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVErandom.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVErandompatch.h
The-Chinese-University-of-Hong-Ko…/.../private
ARCHIVErapidxml.h
The-Chinese-University-of-Hong-Kong-Sh…/.../io
ARCHIVEREADME
The-Chinese-University-of-…/.../party-together
ARCHIVEREADME.txt
The-Chinese-Universi…/.../CSC4130-Assignment-3
ARCHIVEREADME.txt
The-Chinese-Universi…/.../DDA2003-Assignment-3
ARCHIVEREADME.txt
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVEREADME.txt
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVErecursion.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEregexpr.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEregression_result_trial_1.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEregression_result_trial_10.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEregression_result_trial_2.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEregression_result_trial_3.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEregression_result_trial_4.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEregression_result_trial_5.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEregression_result_trial_6.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEregression_result_trial_7.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEregression_result_trial_8.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVEregression_result_trial_9.xlsx
The-Chinese-University-of-Hong-Ko…/.../results
ARCHIVERemoveComments.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-1-src
ARCHIVERemoveComments.h
The-Chinese-Univ…/.../CSC3002-Assignment-1-src
ARCHIVEreport.pdf
The-Chinese-U…/.../CSC3050-Project-2-simulator
ARCHIVEreport.pdf
The-Chinese-Univers…/.../CSC3050-Project-4-CPU
ARCHIVEreport.pdf
The-Chinese-U…/.../CSC3050-Project-1-assembler
ARCHIVEreport.pdf
The-Chinese-Uni…/.../CSC3050-Project-3-verilog
ARCHIVEreport.pdf
The-Chinese-University-…/.../CSC4005-Project-4
ARCHIVEreport.pdf
The-Chinese-University-of-…/.../party-together
ARCHIVEreport.pdf
The-Chinese-Universi…/.../CSC3150-Assignment-1
ARCHIVEreport.pdf
The-Chinese-University-…/.../CSC4005-Project-3
ARCHIVEreport.pdf
The-Chinese-University-…/.../CSC4005-Project-2
ARCHIVEreport.pdf
The-Chinese-Universi…/.../CSC3150-Assignment-3
ARCHIVEreport.pdf
The-Chinese-Universi…/.../CSC3150-Assignment-2
ARCHIVEreport.pdf
The-Chinese-University-…/.../CSC4005-Project-1
ARCHIVEreport.pdf
The-Chinese-Universi…/.../CSC3150-Assignment-4
ARCHIVEreportWebVitals.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVErequire.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVErequirements.txt
The…/.../text-based-image-generation-for-cuhksz-buildings
ARCHIVEresult.txt
The-Chinese-Uni…/.../CSC3050-Project-3-verilog
ARCHIVEreversequeue.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-2-src
ARCHIVEreversequeue.h
The-Chinese-Univ…/.../CSC3002-Assignment-2-src
ARCHIVErun.sh
The-Chinese-University-…/.../CSC4005-Project-3
ARCHIVErun.sh
The-Chinese-University-…/.../CSC4005-Project-4
ARCHIVErun.sh
The-Chinese-University-of-Hong-K…/.../project1
ARCHIVErun.sh
The-Chinese-University-of-Hong-K…/.../project2
ARCHIVEsales-data-set.csv
The-Chinese-University-of-Hong-Ko…/.../data-Q1
ARCHIVEsbatch_PartA.sh
The-Chinese-University-of-Hong-Kong-…/.../Gray
ARCHIVEsbatch_PartA.sh
The-Chinese-University-of-Hong-Ko…/.../scripts
ARCHIVEsbatch_PartB.sh
The-Chinese-University-of-Hong…/.../20K-smooth
ARCHIVEsbatch_PartB.sh
The-Chinese-University-of-Hon…/.../Lena-smooth
ARCHIVEsbatch_PartB.sh
The-Chinese-University-of-Hong-Ko…/.../scripts
ARCHIVEsbatch-bonus.sh
The-Chinese-University-…/.../CSC4005-Project-4
ARCHIVEsbatch-perf.sh
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEsbatch-perf.sh
The-Chinese-University-…/.../CSC4005-Project-3
ARCHIVEsbatch-profile.sh
The-Chinese-University-…/.../CSC4005-Project-4
ARCHIVEsbatch.sh
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEsbatch.sh
The-Chinese-University-…/.../CSC4005-Project-3
ARCHIVEsbatch.sh
The-Chinese-University-…/.../CSC4005-Project-4
ARCHIVEscatter.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEScatterPlot.js
The-Chinese-University-of-Hong…/.../components
ARCHIVEsegment_fault.c
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVEsequential_PartA.cpp
The-Chinese-University-of-Hong-Kong-S…/.../cpu
ARCHIVEsequential_PartB.cpp
The-Chinese-University-of-Hong-Kong-S…/.../cpu
ARCHIVEsequential.cpp
The-Chinese-University-of-Hong…/.../bucketsort
ARCHIVEsequential.cpp
The-Chinese-University-of-Hong-…/.../mergesort
ARCHIVEsequential.cpp
The-Chinese-University-of-H…/.../odd-even-sort
ARCHIVEsequential.cpp
The-Chinese-University-of-Hong-…/.../quicksort
ARCHIVEserver.h
The-Chinese-University-of-Hong-Kong-Sh…/.../io
ARCHIVEset.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEsetupTests.js
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEshows.txt
The-Chinese-University-of-Hong-Ko…/.../data-Q2
ARCHIVEshuffle.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEsimd_PartA.cpp
The-Chinese-University-of-Hong-Kong-S…/.../cpu
ARCHIVEsimd_PartB.cpp
The-Chinese-University-of-Hong-Kong-S…/.../cpu
ARCHIVEsimd.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEsimpio.h
The-Chinese-University-of-Hong-Kong-Sh…/.../io
ARCHIVEsimple_ml_ext.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEsimple_ml_ext.hpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEsimple_ml_openacc.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEsimple_ml_openacc.hpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEsimplegraph.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-6-src
ARCHIVEsimplegraph.h
The-Chinese-Univ…/.../CSC3002-Assignment-6-src
ARCHIVESimpleTextEditor.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-4-src
ARCHIVESimpleTextEditor.h
The-Chinese-Univ…/.../CSC3002-Assignment-4-src
ARCHIVEslurm.sh
The-Chinese-University-of-Hong-Kong…/.../bonus
ARCHIVEslurm.sh
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVEslurm.sh
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVEsoftmax_classifier_openacc.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEsoftmax_classifier.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEsoftmax_openacc.txt
The-Chinese-University-of-Hong-…/.../profiling
ARCHIVEsound.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEsparsegrid.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEspl.cpp
The-Chinese-University-of-…/.../StanfordCPPLib
ARCHIVESPOJ - ACODE.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - ADAINDEX.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - ADFRUITS.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - AIBOHP.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - ANARC09A.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - BSEARCH1.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - BTCODEJ.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - BYTESM2.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - CHAIN.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - CHOCOLA.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - CHOCOLA.py
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - CLFLARR.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - COINS.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - COT.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - CRAYON.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - CUBNUM.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - DQUERY.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - FARIDA.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - GOT.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - GSS3.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - HISTOGRA.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - HORRIBLE.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - HOTELS.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - IITKWPCI.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - KNAPSACK.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - KQUERY.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - LCA.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - LKS.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - M00PAIR.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - MATSUM.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - MISERMAN.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - MKTHNUM.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - MST.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - QTREE2.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - QUEUEEZ.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - RMQSQ.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - RPLB.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - STACKEZ.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - SUMMUL.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - SUMSUM.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - THRBL.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - UPDATEIT.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESPOJ - YODANESS.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVESTA2001 - Assignment 1 Computer-based Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 1 Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 1.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 10 Computer-based Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 10 Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 10.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 11 Computer-based Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 11 Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 11.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 2 Computer-based Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 2 Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 2.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 3 Computer-based Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 3 Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 3.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 4 Computer-based Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 4 Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 4.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 5 Computer-based Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 5 Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 5.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 6 Computer-based Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 6 Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 6.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 7 Computer-based Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 7 Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 7.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 8 Computer-based Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 8 Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 8.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 9 Computer-based Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 9 Problem.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001 - Assignment 9.pdf
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVESTA2001.grades
The-…/.../STA2001-Probability-and-Statistics-I
ARCHIVEstack.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEstackedBarChart.js
The-Chinese-University-of-Hong-Kong-Sh…/.../js
ARCHIVEstate.pvsm
The-Chinese-Universi…/.../DDA2003-Assignment-6
ARCHIVEstatic.h
The-Chinese-University-of-Hong-Ko…/.../private
ARCHIVEstl.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEstop.c
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVEstores-data-set.csv
The-Chinese-University-of-Hong-Ko…/.../data-Q1
ARCHIVEstreamlined-performance-1024x1024.png
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVEstreamlined-performance-2048x2048.png
The-Chinese-University-of-Hong-K…/.../appendix
ARCHIVEstringmap.cpp
The-Chinese-Univ…/.../CSC3002-Assignment-5-src
ARCHIVEstringmap.h
The-Chinese-Univ…/.../CSC3002-Assignment-5-src
ARCHIVEstringutils.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEstrlib.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEstudent_utils.py
The-Chinese-University-of-…/.../party-together
ARCHIVEstyle.css
The-Chinese-University-of-Hong-Kong-S…/.../css
ARCHIVEstyle.css
The-Chinese-University-of-Hong-Kong-S…/.../css
ARCHIVEstyles.css
The-Chinese-Universi…/.../DDA2003-Assignment-2
ARCHIVEstyles.css
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEstyles.css
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVESVM_linear.txt
The-Chinese-Universi…/.../DDA3020-Assignment-2
ARCHIVESVM_poly2.txt
The-Chinese-Universi…/.../DDA3020-Assignment-2
ARCHIVESVM_poly3.txt
The-Chinese-Universi…/.../DDA3020-Assignment-2
ARCHIVESVM_rbf.txt
The-Chinese-Universi…/.../DDA3020-Assignment-2
ARCHIVESVM_sigmoid.txt
The-Chinese-Universi…/.../DDA3020-Assignment-2
ARCHIVESVM_slack.txt
The-Chinese-Universi…/.../DDA3020-Assignment-2
ARCHIVEterminate.c
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVEterrible.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEtest_ALU.v
The-Chinese-Uni…/.../CSC3050-Project-3-verilog
ARCHIVEtest_ALU.vvp
The-Chinese-Uni…/.../CSC3050-Project-3-verilog
ARCHIVEtest_CPU.v
The-Chinese-Univers…/.../CSC3050-Project-4-CPU
ARCHIVEtest.c
The-Chinese-University-of-Hong-K…/.../program2
ARCHIVEtest.sh
The-Chinese-University-…/.../CSC4005-Project-4
ARCHIVEtester.cpp
The-Chinese-U…/.../CSC3050-Project-1-assembler
ARCHIVEtester.h
The-Chinese-U…/.../CSC3050-Project-1-assembler
ARCHIVEtimer.h
The-Chinese-University-of-Hong-Kong-…/.../util
ARCHIVEtokenscanner.h
The-Chinese-University-of-Hong-Kong-Sh…/.../io
ARCHIVEtraining.ipynb
The…/.../text-based-image-generation-for-cuhksz-buildings
ARCHIVEtrap.c
The-Chinese-University-of-Hong-K…/.../program1
ARCHIVEurlstream.h
The-Chinese-University-of-Hong-Kong-Sh…/.../io
ARCHIVEusaco09marsilver3.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEusaco11febsilver2.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEusaco11novsilver2.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEusaco12febgp1.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEusaco12novsilver1.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEusaco13jangold2.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEusaco13opensilver1.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEusaco14decsilver1.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEusaco15decsilver2.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEusaco15febsilver1.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEusaco15febsilver3.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEusaco17febgold1.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEusaco17opensilver1.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEusaco19jansilver3.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEusaco20opensilver1.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEusaco22febsilver2.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEusaco23opengold2.cpp
The-Chinese-University-of-Hong-Kong…/.../SchOJ
ARCHIVEuser_program.cu
The-Chinese-University-of-Hong-Kong…/.../bonus
ARCHIVEuser_program.cu
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVEuser_program.cu
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVEuser-shows.txt
The-Chinese-University-of-Hong-Ko…/.../data-Q2
ARCHIVEutil.c
The-Chinese-University-of-Hon…/.../thread_pool
ARCHIVEutil.h
The-Chinese-University-of-Hon…/.../thread_pool
ARCHIVEutil.o
The-Chinese-University-of-Hon…/.../thread_pool
ARCHIVEutils.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEutils.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEutils.hpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEutils.hpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEutils.py
The-Chinese-University-of-…/.../party-together
ARCHIVEutils.py
The…/.../text-based-image-generation-for-cuhksz-buildings
ARCHIVEutlist.h
The-Chinese-University-of-Hon…/.../thread_pool
ARCHIVEUVA - 10003.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10034.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10139.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10183.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10229.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10271.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10338.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10583.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10783.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10810.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10815.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10819.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10924.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10943.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10943.py
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10946.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10954.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 10977.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 11192.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 11401.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 1149.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 11517.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 11624.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 11715.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 11730.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 11876.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 1197.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 11974.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 11995.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 1230.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 1231.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 1234.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 12470.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 13116.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 138.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 1584.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 160.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 336.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 355.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 378.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 429.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 439.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 484.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 496.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 497.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 501.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 514.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 532.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 571.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 572.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 612.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 621.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 674.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 793.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 920.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 924.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 929.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 957.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEUVA - 989.cpp
Competitive-Progr…/.../Competitive-Programming
ARCHIVEvalidator
The-Chinese-University-of-Hong-K…/.../project2
ARCHIVEvalidator.cpp
The-Chinese-University-of-Hong-Kong-S…/.../src
ARCHIVEvector.h
The-Chinese-University-of-Hon…/.../collections
ARCHIVEversion.h
The-Chinese-University-of-Hong-Ko…/.../private
ARCHIVEvirtual_memory.cu
The-Chinese-University-of-Hong-Kong…/.../bonus
ARCHIVEvirtual_memory.cu
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVEvirtual_memory.h
The-Chinese-University-of-Hong-Kong…/.../bonus
ARCHIVEvirtual_memory.h
The-Chinese-University-of-Hong-Kon…/.../source
ARCHIVEvolume_rendering_with_outline.png
The-Chinese-Universi…/.../DDA2003-Assignment-6
ARCHIVEworld-geojson.json
The-Chinese-University-of-Hong-Kong-…/.../data
ARCHIVExmlutils.h
The-Chinese-University-of-Hong-Kong-Sh…/.../io
[Tutorial] Principal Component Analysis
October 10, 2023 by Yohandi
In data science, Principal Component Analysis (PCA) is a popular technique to reduce the dimension of a dataset. Picture this: data can sometimes be overwhelming with its many dimensions, much like a city with a lot of streets and alleys. In this analogy, PCA doesn't just show us every winding alley – it points us to the main roads and iconic landmarks.
In a more technical term, PCA transforms the data into a set of new variables known as principal components. Each of these components is ranked based on its significance, meaning the most impactful ones capture the majority of the data's variance. As a result, patterns, trends, and outliers become more evident or even discovered in the reduced data; therefore, one can simply focus on these main components.
Principal Component Analysis
The primary objective of PCA is to reduce the complexity of data while preserving as much information as possible. So, instead of dealing with, let's say, 100 different factors, we can just look at the main 2 or 3 that capture most of the story. A simple analogy can be found when recognizing a face: "I know that all faces have similar structures, but this uniqueness of nose and eyebrow combo is John's, for sure!"
The concepts and derivations presented in the following are inspired by and adapted from a session on Principal Component Analysis that I attended in 2022, delivered by Prof. Baoyuan Wu. Any interpretations, extensions, or errors are my own.
There are two objectives for PCA that we are after. First, when reducing the dimension of a dataset, we want to make variance as large as possible. This concept is to spread the data points as widely as possible while keeping their relative positions to each other, pertaining its similarity when reduced. Second, we want to make the reconstruction error as minimum as possible, making sure that we don't lose too much original information. Both ideas' derivation are shown below.
Derivation by Maximal Variance
As an conceptual overview, let's first understand the main reason of having the variance to be maximal. For instance, what does it mean to maximize it?
When we are reducing the dimensions of data using PCA, we are essentially trying to condense the information. By maximizing variance, we ensure that we are focusing on the most crucial patterns or structures in the data. By focusing on high variance, PCA gives us a summary of our data.
Suppose we have a dataset
, where
- KaTeX can only parse string typed expression,
- KaTeX can only parse string typed expression, and
- KaTeX can only parse string typed expression
Denote
This implies that:
Derivation by Minimal Reconstruction Error
Again, as an conceptual overview, let's first understand the main reason of having the reconstruction error to be minimal.
When we project our data onto a lower-dimensional subspace, we are essentially compressing the data, resulting in some information loss. The reconstruction error measures this loss: the difference between the original data and the data reconstructed from its lower-dimensional representation. Ideally, we want this error to be as small as possible, implying our compressed representation is a good approximation of the original data.
By definition, our objective is
Equivalence of Both Derivations
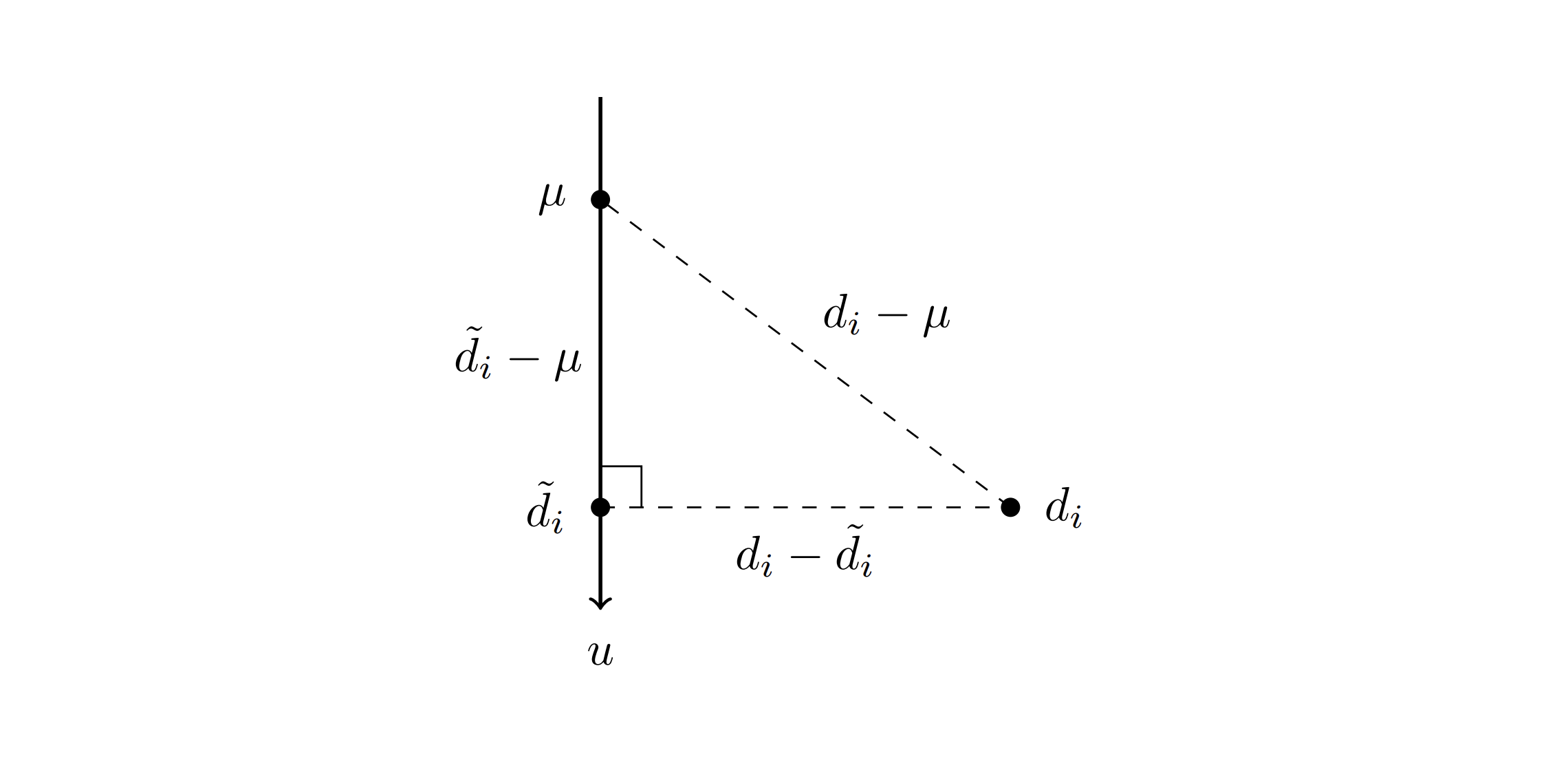
By Pythagorean Theorem,
Since
Lagrange Function Formulation
I touched upon Lagrange Relaxation in a previous blog post, which you can find here. If this topic interests you, I recommend starting there for some foundational understanding.
, where
, where
Singular Value Decomposition
If you are unfamiliar with Singular Value Decomposition, I recommend referring to some resources, one of which is this very blog.
The previous suggests that the optimal solution
, where
Implementation
import numpy as np from numpy.linalg import eig def PCA(D, K): n = len(D) D = np.transpose(np.array(D)) mu = np.mean(D, axis = 1)[:, None] D_mu = D - mu # Calculate empirical covariance matrix empirical_covariance = np.dot(D_mu, np.transpose(D_mu)) / n # Perform SVD decomposition for empirical covariance matrix eigenvalues, eigenvectors = eig(empirical_covariance) # Take the top K eigenvectors sorted_indices = np.argsort(eigenvalues)[::-1] U = eigenvectors[:, sorted_indices[:K]] return np.transpose(np.matmul(np.transpose(U), D - mu)).tolist() D = [ [-1, 2, -3], [2, 0, -1], [1, 1, 1], [-1, -2, 0], [2, 1, 3], [0, -1, 2], [-2, 1, -1], [1, -2, 2], [3, 0, -2], [0, 1, 1] ] print(PCA(D, 1))
Closing
We have seen Principal Component Analysis' ability to simplify complex data. By focusing on what's essential and reducing unnecessary noise, PCA provides a clearer perspective on the underlying patterns in our data. It's a key tool in data science, but it's important to use it wisely, ensuring we don't overlook critical details in our simplicity quest.
References
- Baoyuan Wu, "Principal Component Analysis," The Chinese University of Hong Kong, Shenzhen, 2022.
- Hervé Abdi, Lynne J. Williams, "Principal component analysis," Wiley Interdisciplinary Reviews: Computational Statistics, 2010. https://doi.org/10.1002/wics.101.
© 2023 Yohandi. All rights reserved.